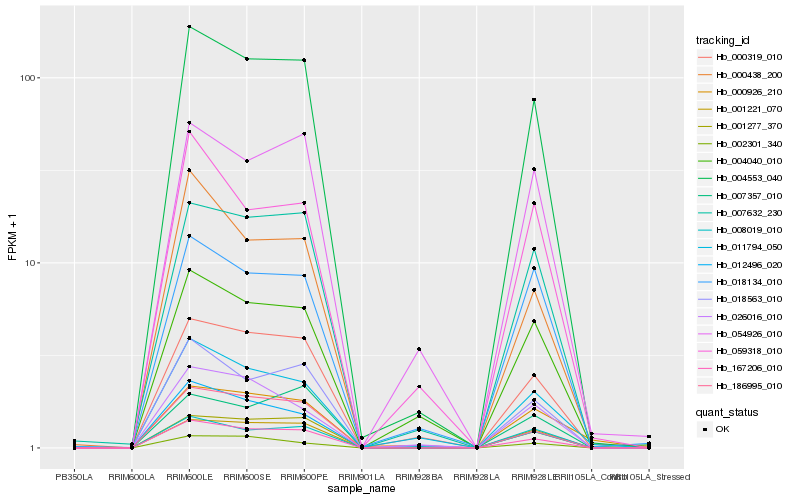
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004553_040 |
0.0 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 2 |
Hb_167206_010 |
0.0520128083 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 3 |
Hb_001277_370 |
0.0651841411 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF30, putative [Ricinus communis] |
| 4 |
Hb_001221_070 |
0.0683785027 |
- |
- |
PREDICTED: calcium-binding protein CML38-like [Jatropha curcas] |
| 5 |
Hb_007632_230 |
0.069556041 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 6 |
Hb_000319_010 |
0.0752945437 |
- |
- |
hypothetical protein POPTR_0016s03600g [Populus trichocarpa] |
| 7 |
Hb_018563_010 |
0.080772995 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 8 |
Hb_054926_010 |
0.1027110966 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 9 |
Hb_008019_010 |
0.1029041242 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 10 |
Hb_186995_010 |
0.1035458081 |
- |
- |
hypothetical protein POPTR_0012s01755g [Populus trichocarpa] |
| 11 |
Hb_004040_010 |
0.1049592307 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 12 |
Hb_018134_010 |
0.106886533 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007357_010 |
0.1166061086 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_000438_200 |
0.1184488733 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |
| 15 |
Hb_011794_050 |
0.1249469352 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_012496_020 |
0.1261693892 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 17 |
Hb_026016_010 |
0.1293673399 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_059318_010 |
0.1297061693 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 19 |
Hb_000926_210 |
0.1307642136 |
- |
- |
LIM and calponin domains-containing protein 1, putative [Theobroma cacao] |
| 20 |
Hb_002301_340 |
0.1310838097 |
transcription factor |
TF Family: B3 |
leafy cotyledon 2 [Ricinus communis] |