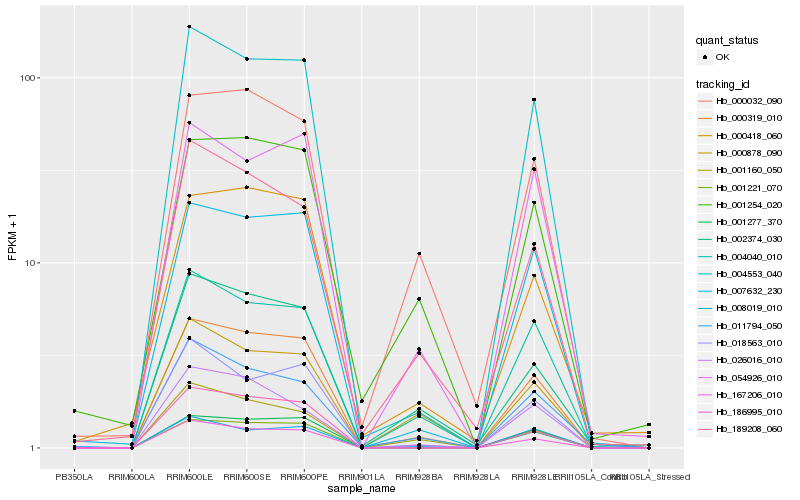
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000319_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s03600g [Populus trichocarpa] |
| 2 |
Hb_004553_040 |
0.0752945437 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 3 |
Hb_007632_230 |
0.0774287539 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 4 |
Hb_004040_010 |
0.0877428974 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 5 |
Hb_001277_370 |
0.0886314911 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF30, putative [Ricinus communis] |
| 6 |
Hb_167206_010 |
0.0894087015 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 7 |
Hb_002374_030 |
0.0939292143 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 8 |
Hb_054926_010 |
0.0981743861 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 9 |
Hb_018563_010 |
0.1006484908 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 10 |
Hb_001221_070 |
0.1023113877 |
- |
- |
PREDICTED: calcium-binding protein CML38-like [Jatropha curcas] |
| 11 |
Hb_000032_090 |
0.1053089166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_186995_010 |
0.1061257556 |
- |
- |
hypothetical protein POPTR_0012s01755g [Populus trichocarpa] |
| 13 |
Hb_000418_060 |
0.1090562381 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 14 |
Hb_189208_060 |
0.1104116221 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 15 |
Hb_001254_020 |
0.1124091778 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 16 |
Hb_000878_090 |
0.1131271643 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 17 |
Hb_008019_010 |
0.1134390951 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 18 |
Hb_011794_050 |
0.1154849922 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_001160_050 |
0.1156292514 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 20 |
Hb_026016_010 |
0.125730127 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |