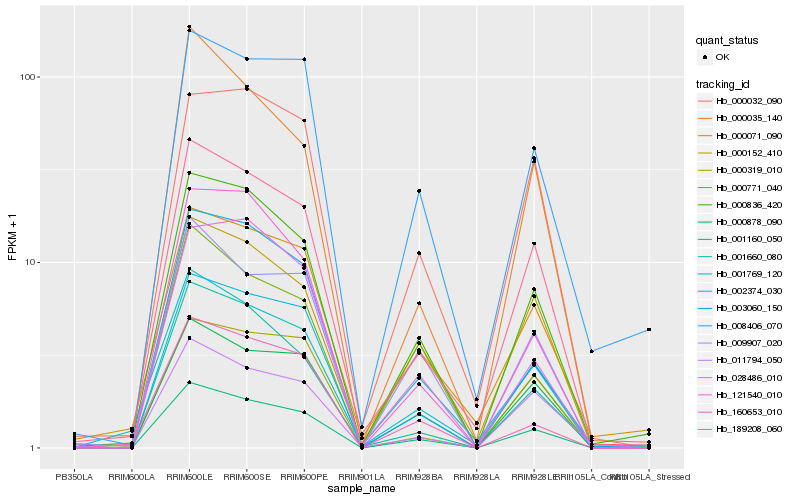
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001160_050 |
0.0 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 2 |
Hb_000836_420 |
0.0672624632 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_002374_030 |
0.0739294949 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000878_090 |
0.0763574668 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 5 |
Hb_189208_060 |
0.078020105 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 6 |
Hb_009907_020 |
0.0907488376 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 7 |
Hb_000152_410 |
0.1001305161 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 8 |
Hb_001660_080 |
0.1083980481 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 9 |
Hb_160653_010 |
0.1103841907 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 10 |
Hb_121540_010 |
0.1118010404 |
- |
- |
PREDICTED: UDP-glycosyltransferase 88A1-like [Jatropha curcas] |
| 11 |
Hb_008406_070 |
0.1144118482 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 12 |
Hb_011794_050 |
0.1146867644 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000319_010 |
0.1156292514 |
- |
- |
hypothetical protein POPTR_0016s03600g [Populus trichocarpa] |
| 14 |
Hb_003060_150 |
0.1170166145 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001769_120 |
0.1179074311 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 16 |
Hb_000771_040 |
0.1182757476 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 17 |
Hb_028486_010 |
0.1183314191 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 18 |
Hb_000071_090 |
0.120789349 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 19 |
Hb_000035_140 |
0.1210638422 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 20 |
Hb_000032_090 |
0.1246629081 |
- |
- |
conserved hypothetical protein [Ricinus communis] |