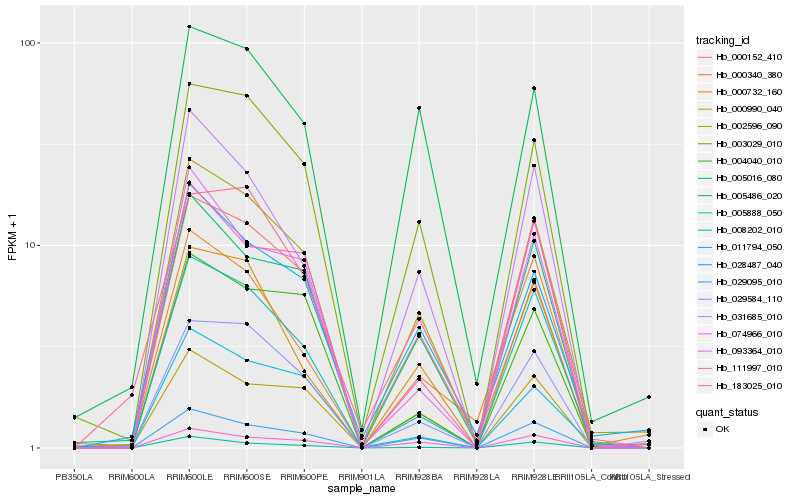
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002596_090 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003029_010 |
0.0827528342 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_028487_040 |
0.0865538427 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |
| 4 |
Hb_005486_020 |
0.0882702185 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 5 |
Hb_029095_010 |
0.0925571281 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 6 |
Hb_000152_410 |
0.0925596997 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 7 |
Hb_029584_110 |
0.0980598153 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 8 |
Hb_008202_010 |
0.1026602922 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_031685_010 |
0.1132830651 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_093364_010 |
0.1140899619 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 11 |
Hb_000990_040 |
0.1199706359 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 12 |
Hb_111997_010 |
0.1256623979 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 13 |
Hb_005016_080 |
0.1257051313 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 14 |
Hb_004040_010 |
0.1259070045 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 15 |
Hb_000732_160 |
0.1295877068 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 16 |
Hb_011794_050 |
0.1298027407 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_074966_010 |
0.1303534801 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 18 |
Hb_000340_380 |
0.1338678247 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 19 |
Hb_005888_050 |
0.1342869919 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_183025_010 |
0.13497297 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |