| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
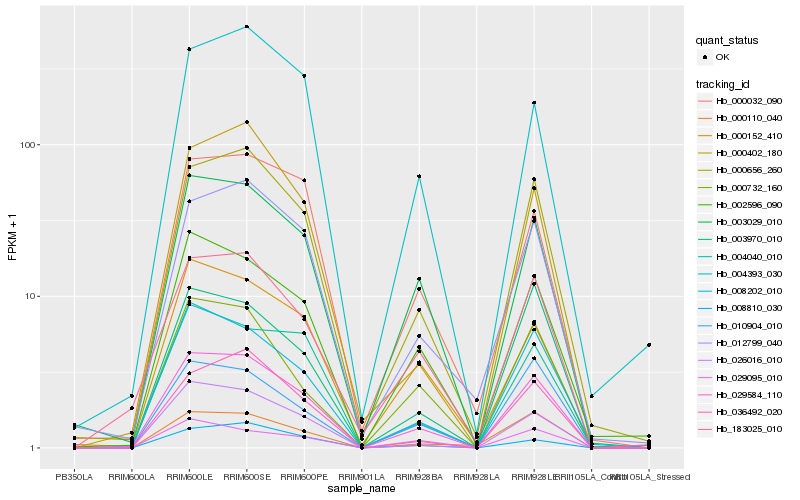
Hb_029584_110 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 2 |
Hb_000656_260 |
0.0807344279 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 3 |
Hb_003029_010 |
0.0813381788 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008202_010 |
0.0907060291 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002596_090 |
0.0980598153 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_183025_010 |
0.1020142031 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 7 |
Hb_026016_010 |
0.1044551168 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_012799_040 |
0.1102384704 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 9 |
Hb_000152_410 |
0.1190447135 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 10 |
Hb_036492_020 |
0.1194814455 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 11 |
Hb_000732_160 |
0.1252864868 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 12 |
Hb_008810_030 |
0.1275225885 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000032_090 |
0.127960869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004040_010 |
0.131178424 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 15 |
Hb_000402_180 |
0.1316237363 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 16 |
Hb_000110_040 |
0.1320641318 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 17 |
Hb_004393_030 |
0.1327811621 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 18 |
Hb_010904_010 |
0.1329377249 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 19 |
Hb_029095_010 |
0.1333703155 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 20 |
Hb_003970_010 |
0.137463025 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |