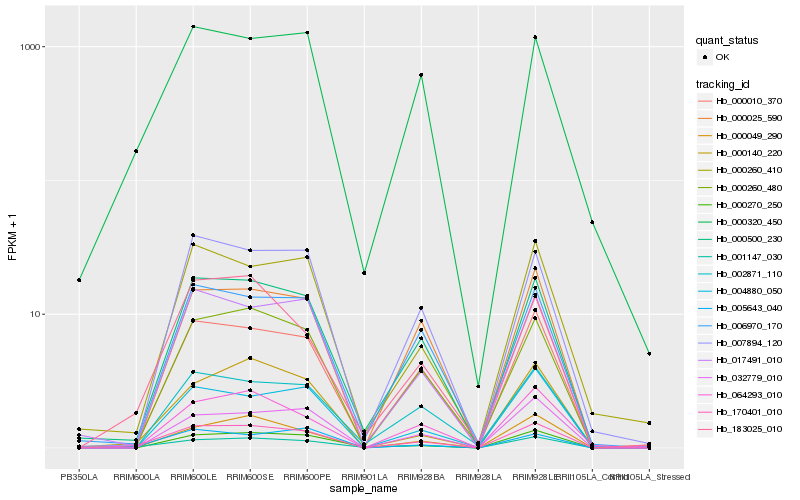
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_170401_010 |
0.0 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 2 |
Hb_001147_030 |
0.0859226556 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 3 |
Hb_000500_230 |
0.1097178636 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 4 |
Hb_000010_370 |
0.112727484 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 5 |
Hb_000260_480 |
0.1142383702 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 6 |
Hb_000140_220 |
0.1320091654 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 7 |
Hb_000049_290 |
0.1320483981 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_183025_010 |
0.1328511474 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 9 |
Hb_017491_010 |
0.1336339541 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007894_120 |
0.1369234273 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_032779_010 |
0.1395877101 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000025_590 |
0.140151864 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_006970_170 |
0.1404743061 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000260_410 |
0.144533051 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 15 |
Hb_000270_250 |
0.1449787744 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s07900g [Populus trichocarpa] |
| 16 |
Hb_000320_450 |
0.1454538945 |
- |
- |
secretory peroxidase [Hevea brasiliensis] |
| 17 |
Hb_002871_110 |
0.1468072398 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 18 |
Hb_004880_050 |
0.1472340287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005643_040 |
0.1494093983 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_064293_010 |
0.1496241843 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |