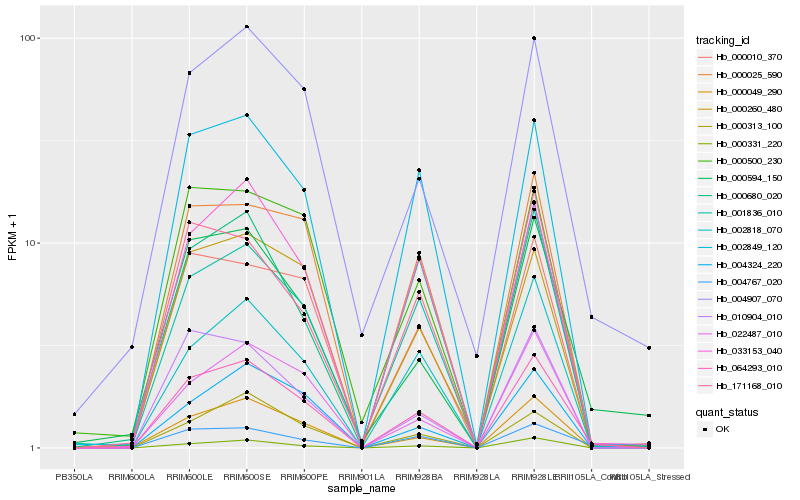
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_064293_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_033153_040 |
0.1023149345 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_002849_120 |
0.1064970892 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 4 |
Hb_000049_290 |
0.1105592285 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_004767_020 |
0.1115774993 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_000331_220 |
0.1125408529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000025_590 |
0.1139753507 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001836_010 |
0.1180125553 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 9 |
Hb_002818_070 |
0.1181879311 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000010_370 |
0.1229753859 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 11 |
Hb_171168_010 |
0.1240559066 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000260_480 |
0.1243121017 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 13 |
Hb_022487_010 |
0.1253475899 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 14 |
Hb_004324_220 |
0.1260585 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 15 |
Hb_010904_010 |
0.1291644037 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000594_150 |
0.1293969183 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 17 |
Hb_000313_100 |
0.1313974992 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 18 |
Hb_000680_020 |
0.1327193961 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000500_230 |
0.1344655491 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 20 |
Hb_004907_070 |
0.1347068985 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |