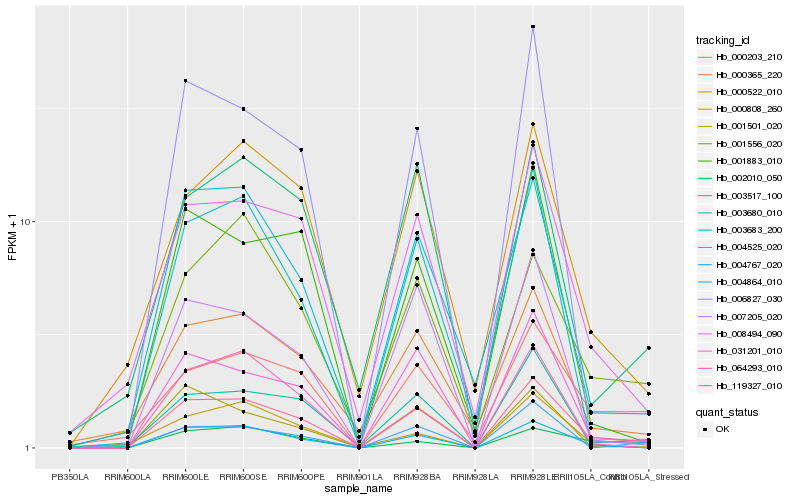
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_119327_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 2 |
Hb_031201_010 |
0.1125813323 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_004767_020 |
0.1227590962 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 4 |
Hb_008494_090 |
0.1421729192 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 5 |
Hb_000203_210 |
0.1484528926 |
- |
- |
PREDICTED: uncharacterized protein LOC105040619 isoform X3 [Elaeis guineensis] |
| 6 |
Hb_004864_010 |
0.153168209 |
- |
- |
hypothetical protein CICLE_v10003960mg, partial [Citrus clementina] |
| 7 |
Hb_007205_020 |
0.1533604017 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 8 |
Hb_004525_020 |
0.1552023273 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 9 |
Hb_000365_220 |
0.1597960471 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 10 |
Hb_003680_010 |
0.1607386339 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_003517_100 |
0.1623336115 |
transcription factor |
TF Family: GRAS |
- |
| 12 |
Hb_000522_010 |
0.1630285059 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 13 |
Hb_001883_010 |
0.1666742483 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 14 |
Hb_001501_020 |
0.1671453871 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 15 |
Hb_003683_200 |
0.1677329031 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 16 |
Hb_006827_030 |
0.1700702806 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 17 |
Hb_001556_020 |
0.1703752614 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 18 |
Hb_000808_260 |
0.1736624759 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 19 |
Hb_064293_010 |
0.1739999305 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_002010_050 |
0.1763629605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |