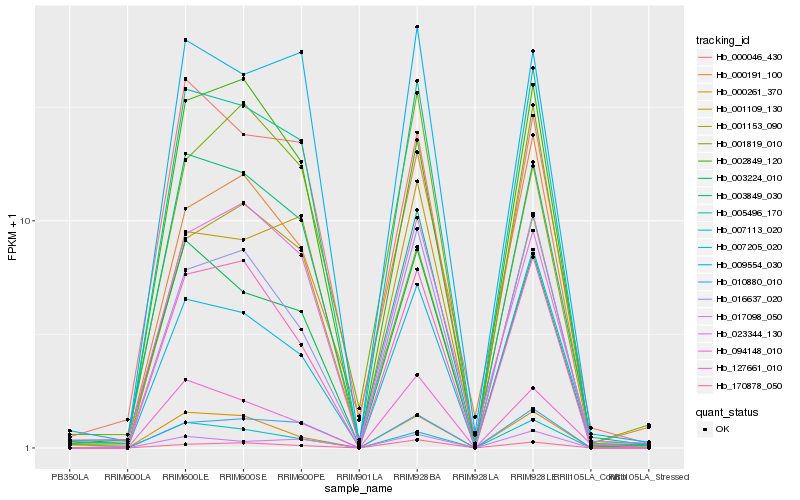
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005496_170 |
0.0 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 2 |
Hb_010880_010 |
0.0724799243 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_003224_010 |
0.081270419 |
- |
- |
- |
| 4 |
Hb_127661_010 |
0.0902023911 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_017098_050 |
0.0951186045 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 6 |
Hb_000261_370 |
0.0998473546 |
- |
- |
- |
| 7 |
Hb_016637_020 |
0.1012198173 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 8 |
Hb_009554_030 |
0.1016312509 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 9 |
Hb_007205_020 |
0.1021103548 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 10 |
Hb_007113_020 |
0.1039377571 |
- |
- |
- |
| 11 |
Hb_001109_130 |
0.1080271825 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003849_030 |
0.1111107985 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 13 |
Hb_001819_010 |
0.11939949 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 14 |
Hb_001153_090 |
0.1197150254 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 15 |
Hb_023344_130 |
0.1201583382 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_000046_430 |
0.1211286256 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 17 |
Hb_170878_050 |
0.1214781854 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 18 |
Hb_002849_120 |
0.1216206285 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 19 |
Hb_000191_100 |
0.1223581435 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_094148_010 |
0.123561683 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |