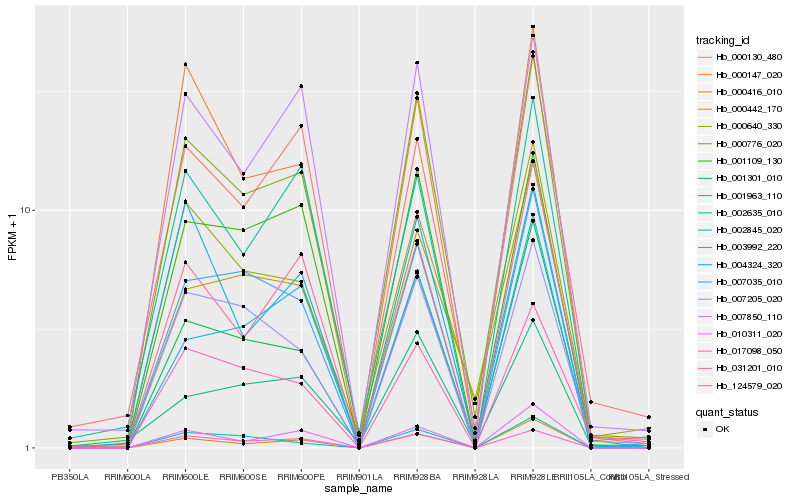
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000776_020 |
0.0 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 2 |
Hb_001301_010 |
0.0754100407 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_007850_110 |
0.1036513256 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 4 |
Hb_000416_010 |
0.1045718347 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 5 |
Hb_017098_050 |
0.1046767429 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 6 |
Hb_000442_170 |
0.1068979415 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 7 |
Hb_001109_130 |
0.1132684309 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007205_020 |
0.1156947746 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 9 |
Hb_124579_020 |
0.1157961992 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 10 |
Hb_010311_020 |
0.116658261 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000147_020 |
0.1222267521 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 12 |
Hb_031201_010 |
0.1269527528 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_002635_010 |
0.1273014738 |
- |
- |
homogentisate phytyl transferase [Hevea brasiliensis] |
| 14 |
Hb_000130_480 |
0.127425942 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 15 |
Hb_000640_330 |
0.1275669805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002845_020 |
0.127734378 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 17 |
Hb_003992_220 |
0.1281863861 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 18 |
Hb_004324_320 |
0.1315079559 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 19 |
Hb_001963_110 |
0.1324386021 |
- |
- |
- |
| 20 |
Hb_007035_010 |
0.1372476695 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |