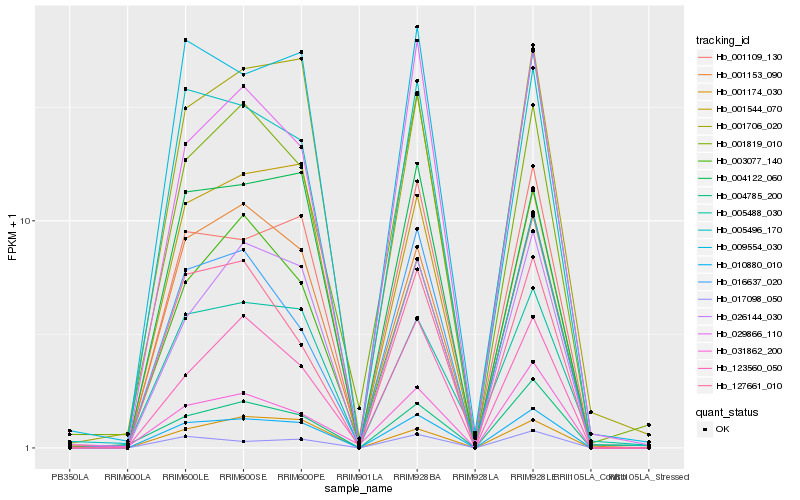
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010880_010 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_005496_170 |
0.0724799243 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 3 |
Hb_001109_130 |
0.0813065082 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004122_060 |
0.0971173397 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 5 |
Hb_026144_030 |
0.0988020543 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 6 |
Hb_017098_050 |
0.1040374741 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 7 |
Hb_001153_090 |
0.1056193949 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 8 |
Hb_001706_020 |
0.1064325262 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 9 |
Hb_009554_030 |
0.1084116636 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 10 |
Hb_001819_010 |
0.108416006 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 11 |
Hb_004785_200 |
0.1098710187 |
- |
- |
PREDICTED: protein MKS1 [Jatropha curcas] |
| 12 |
Hb_005488_030 |
0.1112276119 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 13 |
Hb_123560_050 |
0.1179044689 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 14 |
Hb_003077_140 |
0.1196595114 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 15 |
Hb_001174_030 |
0.1220239026 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_127661_010 |
0.1221379616 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_016637_020 |
0.1223398883 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 18 |
Hb_031862_200 |
0.1232558559 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 19 |
Hb_001544_070 |
0.1238691064 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 20 |
Hb_029866_110 |
0.1242180313 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |