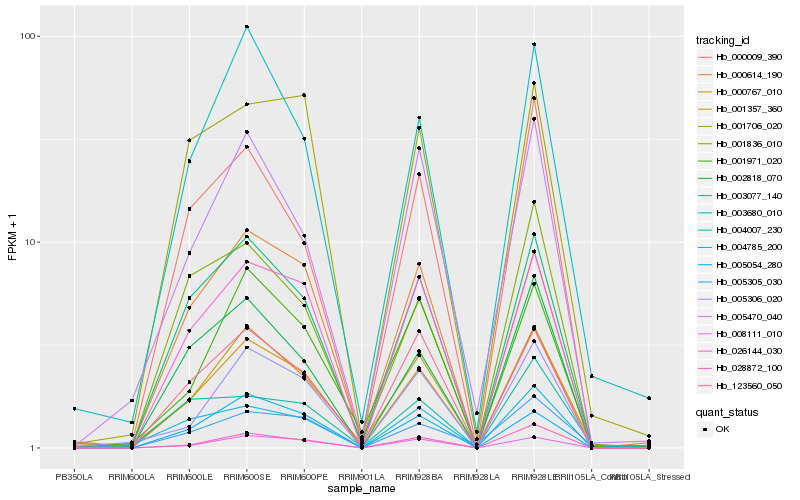
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000767_010 |
0.0 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 2 |
Hb_003077_140 |
0.0880446683 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 3 |
Hb_026144_030 |
0.0896615662 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 4 |
Hb_001357_360 |
0.0989039114 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_028872_100 |
0.1102980052 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 6 |
Hb_008111_010 |
0.1103247108 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 7 |
Hb_004007_230 |
0.1161663508 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002818_070 |
0.1170252116 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_123560_050 |
0.1234947281 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 10 |
Hb_005306_020 |
0.1250271297 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 11 |
Hb_000614_190 |
0.1253353192 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 12 |
Hb_001971_020 |
0.1283121882 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 13 |
Hb_004785_200 |
0.133329646 |
- |
- |
PREDICTED: protein MKS1 [Jatropha curcas] |
| 14 |
Hb_005054_280 |
0.1338424684 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 15 |
Hb_000009_390 |
0.1356737959 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001706_020 |
0.136383569 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 17 |
Hb_005305_030 |
0.1383996106 |
- |
- |
hypothetical protein JCGZ_25922 [Jatropha curcas] |
| 18 |
Hb_005470_040 |
0.1390406091 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 19 |
Hb_001836_010 |
0.1434358183 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 20 |
Hb_003680_010 |
0.1447775795 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |