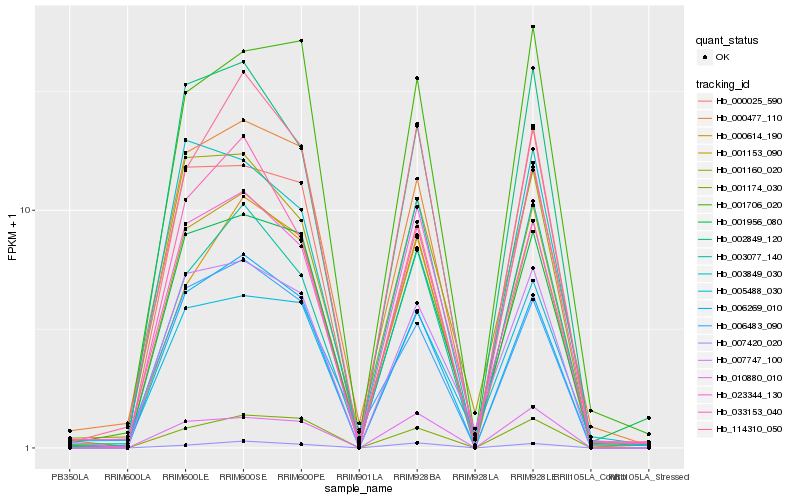
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001153_090 |
0.0 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 2 |
Hb_006269_010 |
0.0735080648 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 3 |
Hb_002849_120 |
0.0752207693 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 4 |
Hb_007747_100 |
0.0821638986 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 5 |
Hb_023344_130 |
0.0834056266 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_001160_020 |
0.0881792844 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 7 |
Hb_003077_140 |
0.0882648715 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 8 |
Hb_001956_080 |
0.0913664183 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 9 |
Hb_001174_030 |
0.0918796756 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000477_110 |
0.0928130845 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 11 |
Hb_033153_040 |
0.0956256061 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_005488_030 |
0.098685543 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 13 |
Hb_114310_050 |
0.1001363332 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 14 |
Hb_003849_030 |
0.1012481208 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 15 |
Hb_006483_090 |
0.1032179003 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 16 |
Hb_000614_190 |
0.1034367185 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 17 |
Hb_010880_010 |
0.1056193949 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001706_020 |
0.1074477301 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 19 |
Hb_007420_020 |
0.1083675662 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 20 |
Hb_000025_590 |
0.1093860068 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |