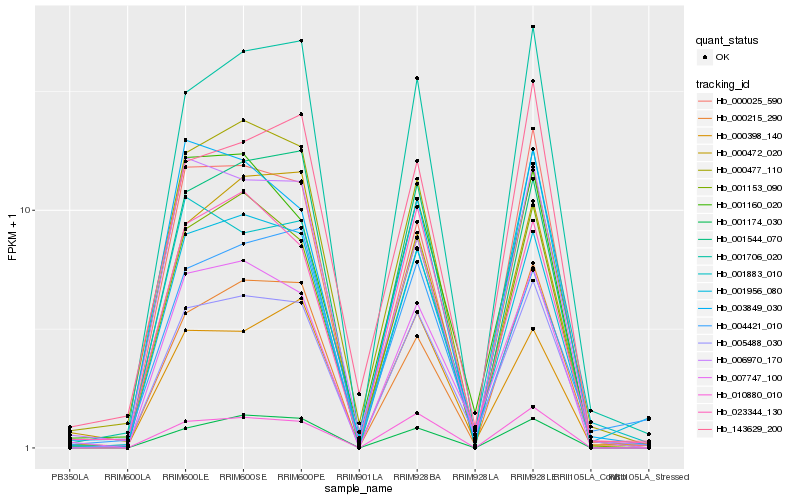
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005488_030 |
0.0 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 2 |
Hb_001706_020 |
0.0850012297 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 3 |
Hb_001956_080 |
0.0882680922 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 4 |
Hb_007747_100 |
0.0957755433 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 5 |
Hb_000215_290 |
0.0972221362 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001153_090 |
0.098685543 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 7 |
Hb_023344_130 |
0.1109754415 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_010880_010 |
0.1112276119 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000025_590 |
0.1155569152 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001174_030 |
0.115829828 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_143629_200 |
0.1180233402 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 12 |
Hb_001544_070 |
0.1212219873 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 13 |
Hb_000477_110 |
0.1215583641 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 14 |
Hb_000472_020 |
0.1217635069 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 15 |
Hb_006970_170 |
0.1229333788 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000398_140 |
0.1231756149 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 17 |
Hb_001160_020 |
0.1257545369 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 18 |
Hb_004421_010 |
0.1258352028 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_001883_010 |
0.1266484975 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 20 |
Hb_003849_030 |
0.1279288456 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |