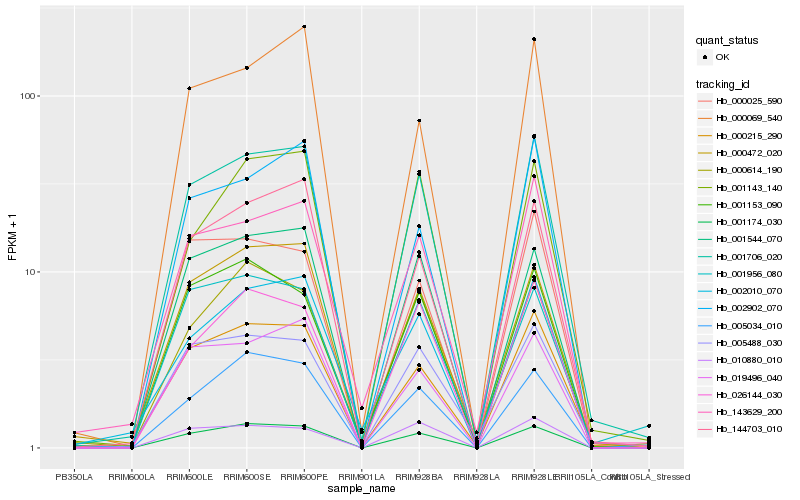
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001706_020 |
0.0 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 2 |
Hb_000215_290 |
0.0695559073 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001174_030 |
0.0740886786 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_005488_030 |
0.0850012297 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 5 |
Hb_143629_200 |
0.0862125034 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 6 |
Hb_001544_070 |
0.0916246412 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 7 |
Hb_000472_020 |
0.0937132089 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 8 |
Hb_144703_010 |
0.0962794468 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 9 |
Hb_026144_030 |
0.1000069995 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 10 |
Hb_019496_040 |
0.1011188556 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 11 |
Hb_002010_070 |
0.1020137924 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 12 |
Hb_001143_140 |
0.1024722253 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_010880_010 |
0.1064325262 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001153_090 |
0.1074477301 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 15 |
Hb_000025_590 |
0.1118555535 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_005034_010 |
0.1124074288 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_001956_080 |
0.1128871809 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 18 |
Hb_000614_190 |
0.113082211 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 19 |
Hb_002902_070 |
0.114672967 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 20 |
Hb_000069_540 |
0.1164180161 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |