| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
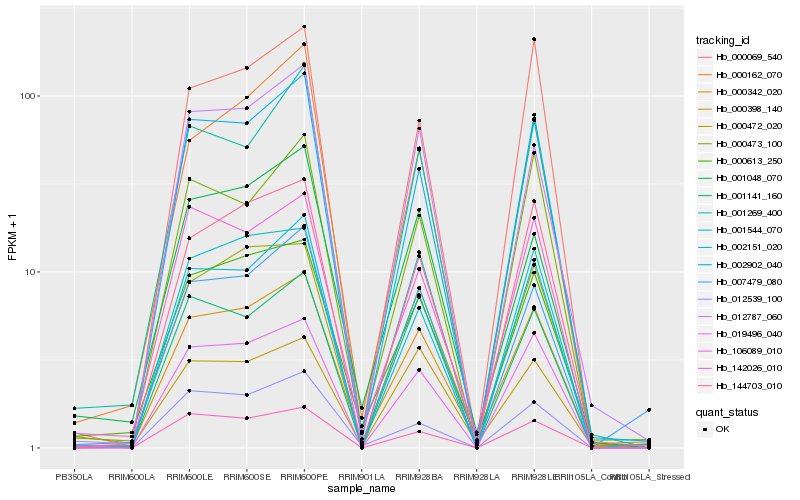
Hb_000342_020 |
0.0 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 2 |
Hb_000613_250 |
0.0745010884 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 3 |
Hb_000472_020 |
0.0803525614 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 4 |
Hb_002902_040 |
0.0804566215 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 5 |
Hb_144703_010 |
0.0811753332 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 6 |
Hb_019496_040 |
0.0832691124 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 7 |
Hb_002151_020 |
0.091171673 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 8 |
Hb_001269_400 |
0.0912656806 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 9 |
Hb_106089_010 |
0.0946242043 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 10 |
Hb_001544_070 |
0.1007405629 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 11 |
Hb_012787_060 |
0.1028839873 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 12 |
Hb_001048_070 |
0.1039581084 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 13 |
Hb_000069_540 |
0.105516029 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 14 |
Hb_000473_100 |
0.1058036528 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 15 |
Hb_007479_080 |
0.1070631001 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 16 |
Hb_142026_010 |
0.1078451209 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 17 |
Hb_000162_070 |
0.1105448138 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 18 |
Hb_000398_140 |
0.1129531317 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 19 |
Hb_001141_160 |
0.1158271383 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 20 |
Hb_012539_100 |
0.1170538777 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK11 [Jatropha curcas] |