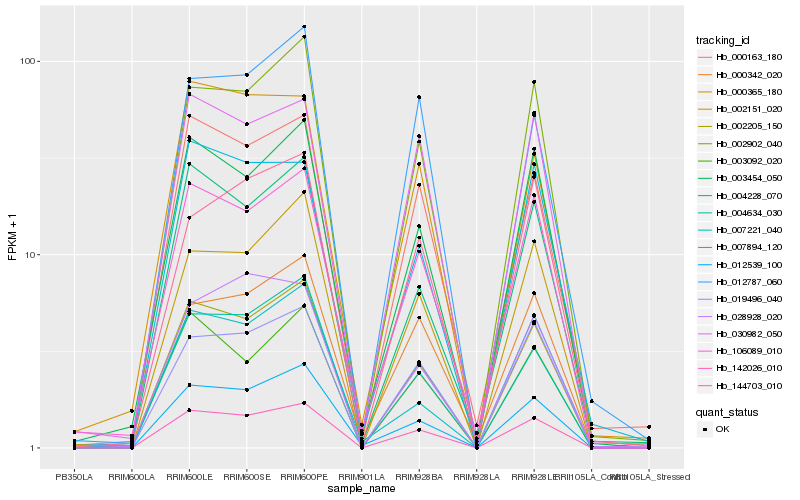
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142026_010 |
0.0 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 2 |
Hb_002205_150 |
0.0580645685 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 3 |
Hb_002902_040 |
0.0679532547 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 4 |
Hb_019496_040 |
0.073528134 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 5 |
Hb_106089_010 |
0.0758110759 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 6 |
Hb_003454_050 |
0.0784372839 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 7 |
Hb_000163_180 |
0.0821042723 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 8 |
Hb_004634_030 |
0.0877732284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002151_020 |
0.0902961299 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 10 |
Hb_030982_050 |
0.0906654192 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 11 |
Hb_004228_070 |
0.0912302046 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 12 |
Hb_012539_100 |
0.0930243538 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK11 [Jatropha curcas] |
| 13 |
Hb_003092_020 |
0.0937480506 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 14 |
Hb_007894_120 |
0.1040915633 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000342_020 |
0.1078451209 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 16 |
Hb_000365_180 |
0.112492605 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 17 |
Hb_012787_060 |
0.1125909401 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 18 |
Hb_144703_010 |
0.1164482791 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 19 |
Hb_028928_020 |
0.1175175434 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 20 |
Hb_007221_040 |
0.1183864395 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |