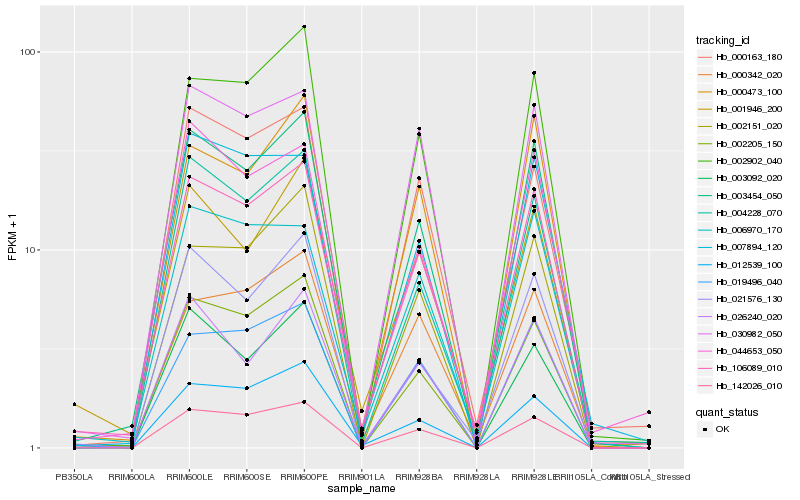
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106089_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 2 |
Hb_003454_050 |
0.0522684505 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 3 |
Hb_142026_010 |
0.0758110759 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 4 |
Hb_002205_150 |
0.0830002395 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 5 |
Hb_030982_050 |
0.0878237749 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 6 |
Hb_002902_040 |
0.0879162293 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 7 |
Hb_006970_170 |
0.0932629775 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_019496_040 |
0.0935031969 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 9 |
Hb_012539_100 |
0.0936967271 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK11 [Jatropha curcas] |
| 10 |
Hb_000342_020 |
0.0946242043 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 11 |
Hb_004228_070 |
0.0975819585 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 12 |
Hb_001946_200 |
0.0981107795 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_000473_100 |
0.100290527 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 14 |
Hb_000163_180 |
0.1009733548 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 15 |
Hb_003092_020 |
0.1043543537 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 16 |
Hb_026240_020 |
0.1051563827 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 17 |
Hb_044653_050 |
0.1070742711 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002151_020 |
0.1072868262 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 19 |
Hb_021576_130 |
0.1082775226 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007894_120 |
0.1091887627 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |