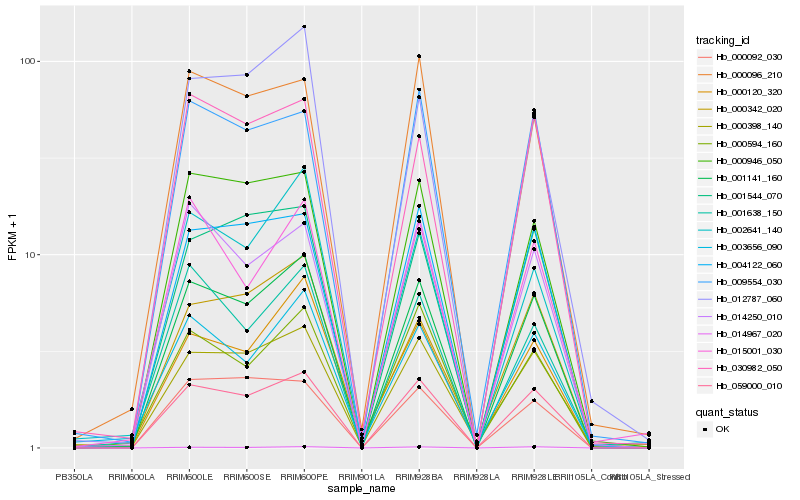
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_160 |
0.0 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 2 |
Hb_059000_010 |
0.0393204364 |
- |
- |
hypothetical protein JCGZ_17736 [Jatropha curcas] |
| 3 |
Hb_004122_060 |
0.0905458214 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 4 |
Hb_000946_050 |
0.0942321944 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_000398_140 |
0.0974256939 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 6 |
Hb_030982_050 |
0.0975195756 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 7 |
Hb_001544_070 |
0.1006873527 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 8 |
Hb_014250_010 |
0.1018049667 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 9 |
Hb_000120_320 |
0.1034145151 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_009554_030 |
0.1050864934 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 11 |
Hb_003656_090 |
0.1056767397 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 12 |
Hb_012787_060 |
0.107551344 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 13 |
Hb_000594_160 |
0.1077970754 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 14 |
Hb_002641_140 |
0.109907694 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_000096_210 |
0.1112000674 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 16 |
Hb_001638_150 |
0.1112553764 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 17 |
Hb_014967_020 |
0.1124801269 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_015001_030 |
0.1138192836 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 19 |
Hb_000342_020 |
0.1158271383 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 20 |
Hb_000092_030 |
0.1161844508 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |