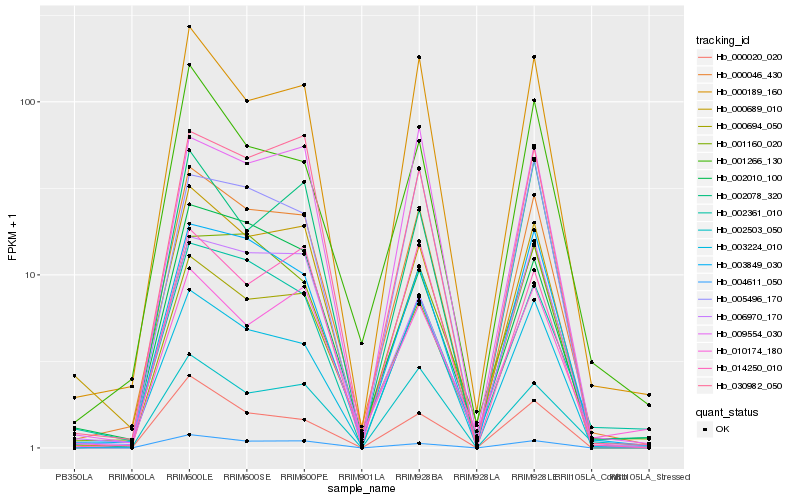
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_430 |
0.0 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 2 |
Hb_000189_160 |
0.0773015587 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 3 |
Hb_003849_030 |
0.0780455882 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 4 |
Hb_003224_010 |
0.0959953246 |
- |
- |
- |
| 5 |
Hb_000689_010 |
0.0966728942 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_030982_050 |
0.0991378722 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 7 |
Hb_004611_050 |
0.1045531753 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 8 |
Hb_006970_170 |
0.1076878626 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_002503_050 |
0.1086813321 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 10 |
Hb_000020_020 |
0.1129068148 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 11 |
Hb_001160_020 |
0.1146306942 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 12 |
Hb_001266_130 |
0.1158953332 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 13 |
Hb_002010_100 |
0.1163498015 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 14 |
Hb_009554_030 |
0.1167397692 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 15 |
Hb_002361_010 |
0.1175636803 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_000694_050 |
0.1184621372 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 17 |
Hb_014250_010 |
0.1193904551 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 18 |
Hb_010174_180 |
0.1195246119 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_002078_320 |
0.1207714959 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_005496_170 |
0.1211286256 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |