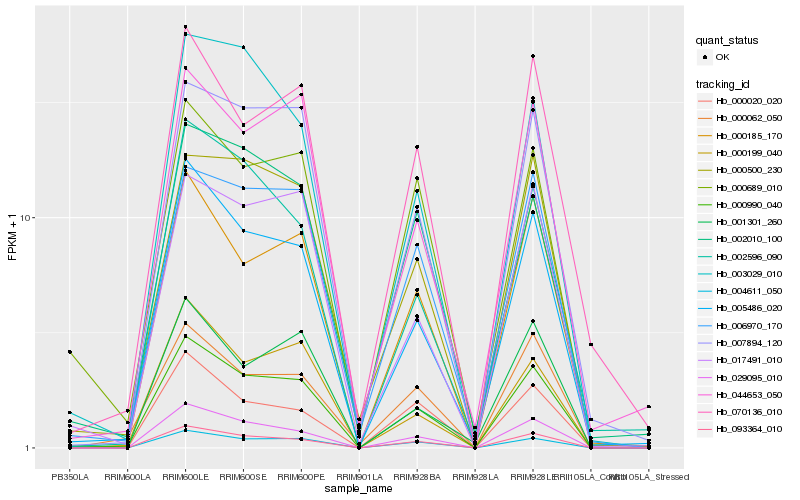
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000990_040 |
0.0 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 2 |
Hb_005486_020 |
0.0665451844 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 3 |
Hb_093364_010 |
0.0724723214 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 4 |
Hb_004611_050 |
0.0744757285 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 5 |
Hb_029095_010 |
0.0797021975 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 6 |
Hb_000185_170 |
0.0973225489 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_007894_120 |
0.1010869236 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_006970_170 |
0.1068398521 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_017491_010 |
0.1073825471 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000689_010 |
0.1084893538 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 11 |
Hb_044653_050 |
0.1098162296 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 12 |
Hb_070136_010 |
0.1121624702 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 13 |
Hb_000500_230 |
0.1127439523 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 14 |
Hb_000199_040 |
0.1158922704 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003029_010 |
0.1190905948 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002596_090 |
0.1199706359 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000062_050 |
0.1218898739 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_001301_260 |
0.1220162446 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000020_020 |
0.1225874289 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 20 |
Hb_002010_100 |
0.1253473864 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |