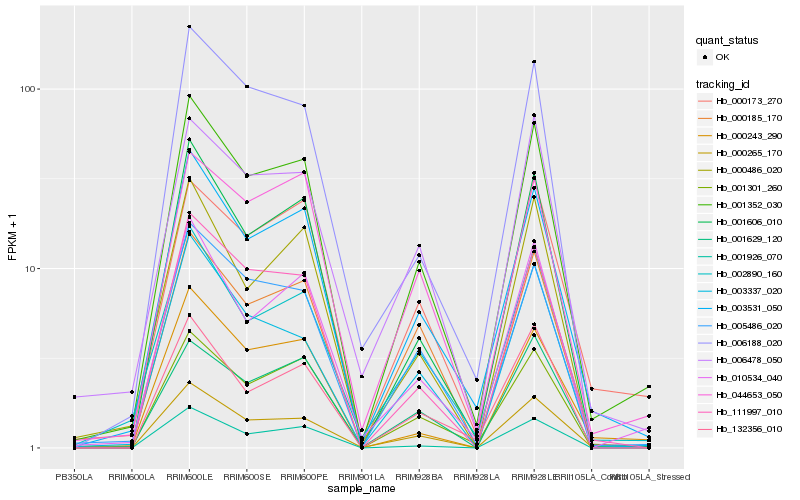
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001352_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 2 |
Hb_003531_050 |
0.06173567 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000265_170 |
0.0787462765 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 4 |
Hb_001606_010 |
0.0812297125 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_010534_040 |
0.0822030516 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005486_020 |
0.0845307831 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 7 |
Hb_002890_160 |
0.0903158485 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001301_260 |
0.0931415169 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_111997_010 |
0.0942192944 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 10 |
Hb_000185_170 |
0.0995833443 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_001629_120 |
0.1033971626 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 12 |
Hb_001926_070 |
0.1055327227 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 13 |
Hb_000173_270 |
0.1073312509 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 14 |
Hb_044653_050 |
0.1075538007 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006478_050 |
0.1077507059 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000486_020 |
0.1079784124 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 17 |
Hb_000243_290 |
0.1083098302 |
- |
- |
plastidic aldolase family protein [Populus trichocarpa] |
| 18 |
Hb_006188_020 |
0.1105664062 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 19 |
Hb_132356_010 |
0.1106515068 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 20 |
Hb_003337_020 |
0.1117998167 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |