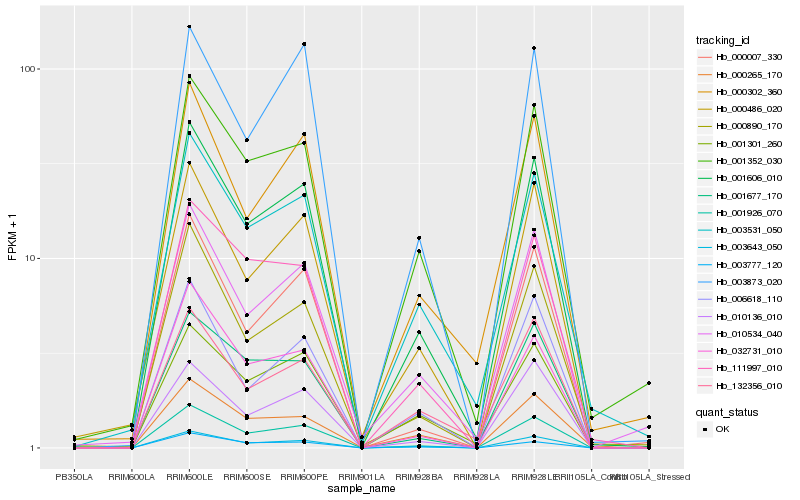
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001606_010 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_001926_070 |
0.0404068422 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 3 |
Hb_000890_170 |
0.0802038176 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 4 |
Hb_001352_030 |
0.0812297125 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 5 |
Hb_000486_020 |
0.0825767131 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 6 |
Hb_032731_010 |
0.0850931485 |
- |
- |
RING-H2 finger protein ATL5M precursor, putative [Ricinus communis] |
| 7 |
Hb_003531_050 |
0.0853844166 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_111997_010 |
0.0867070332 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 9 |
Hb_000007_330 |
0.0871337561 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 10 |
Hb_010534_040 |
0.0877211209 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001301_260 |
0.0883242664 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000302_360 |
0.0889688367 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 13 |
Hb_001677_170 |
0.0955817458 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 14 |
Hb_006618_110 |
0.0959410196 |
- |
- |
- |
| 15 |
Hb_000265_170 |
0.0961320249 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_010136_010 |
0.0969707749 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 17 |
Hb_003643_050 |
0.0974558104 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_003873_020 |
0.1005702389 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 19 |
Hb_003777_120 |
0.1008951632 |
- |
- |
- |
| 20 |
Hb_132356_010 |
0.1021725685 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |