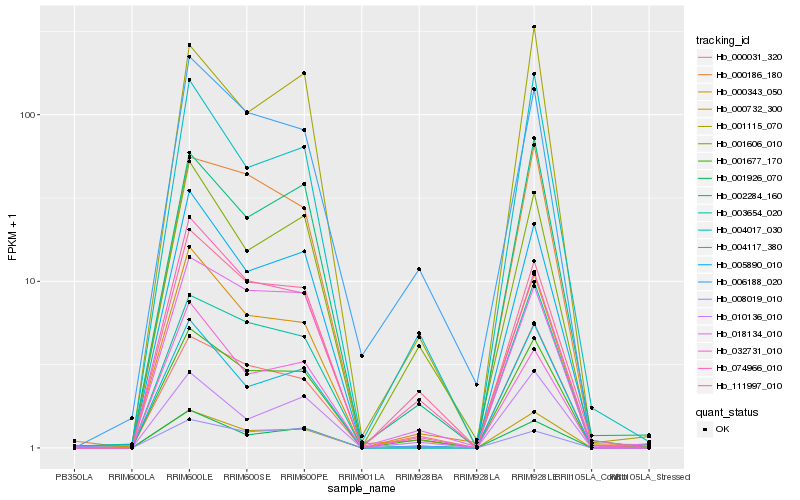
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001677_170 |
0.0 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 2 |
Hb_001926_070 |
0.0756981317 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 3 |
Hb_111997_010 |
0.076682359 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 4 |
Hb_000343_050 |
0.0772240116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000031_320 |
0.0817714707 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_006188_020 |
0.0896775787 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 7 |
Hb_001606_010 |
0.0955817458 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_005890_010 |
0.0981088428 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 9 |
Hb_002284_160 |
0.1006006406 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_004017_030 |
0.1008469923 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 11 |
Hb_018134_010 |
0.1009621955 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010136_010 |
0.1043162234 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 13 |
Hb_003654_020 |
0.1073126087 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_001115_070 |
0.1073584224 |
- |
- |
RecName: Full=CASP-like protein 2C1; Short=PtCASPL2C1 [Populus trichocarpa] |
| 15 |
Hb_004117_380 |
0.1085135928 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 16 |
Hb_000732_300 |
0.1118738632 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000186_180 |
0.1119870054 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 18 |
Hb_074966_010 |
0.1136246433 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 19 |
Hb_008019_010 |
0.1139910018 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 20 |
Hb_032731_010 |
0.1145940587 |
- |
- |
RING-H2 finger protein ATL5M precursor, putative [Ricinus communis] |