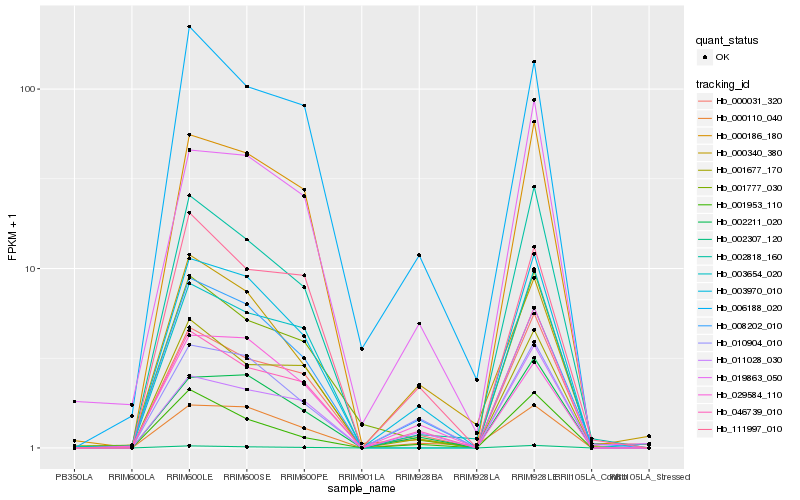
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003970_010 |
0.0 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 2 |
Hb_002211_020 |
0.0613373432 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 3 |
Hb_010904_010 |
0.0700851799 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000031_320 |
0.0865772866 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_008202_010 |
0.098151859 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000110_040 |
0.103876138 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 7 |
Hb_046739_010 |
0.1092193333 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002818_160 |
0.1100181538 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 9 |
Hb_000186_180 |
0.114523412 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 10 |
Hb_001677_170 |
0.1201006995 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 11 |
Hb_011028_030 |
0.1205836125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001953_110 |
0.1254713782 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 13 |
Hb_019863_050 |
0.1310963648 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 14 |
Hb_002307_120 |
0.131993523 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 15 |
Hb_006188_020 |
0.1335278238 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 16 |
Hb_003654_020 |
0.136351747 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_111997_010 |
0.1370151471 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 18 |
Hb_029584_110 |
0.137463025 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 19 |
Hb_001777_030 |
0.1382202847 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 20 |
Hb_000340_380 |
0.139872403 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |