| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
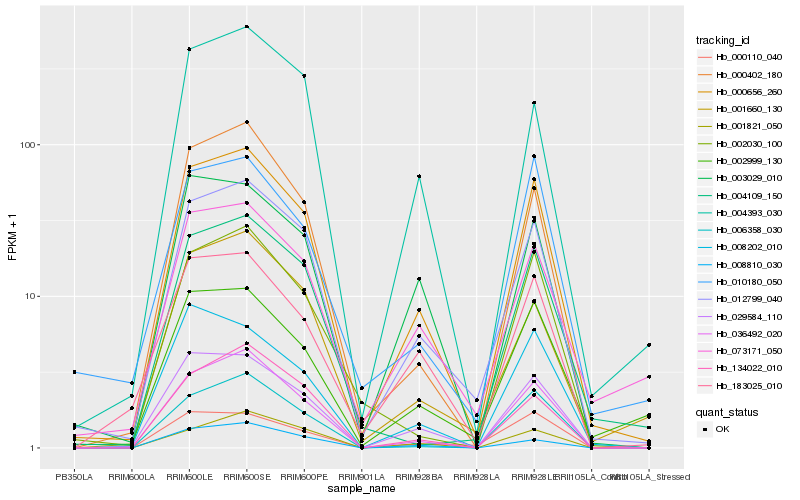
Hb_000656_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 2 |
Hb_012799_040 |
0.0683025691 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 3 |
Hb_036492_020 |
0.0793366725 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 4 |
Hb_029584_110 |
0.0807344279 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 5 |
Hb_006358_030 |
0.1115806521 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 6 |
Hb_000402_180 |
0.1118190744 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 7 |
Hb_002999_130 |
0.1180782596 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 8 |
Hb_183025_010 |
0.1190566407 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 9 |
Hb_003029_010 |
0.1194272897 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002030_100 |
0.1229638337 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 11 |
Hb_073171_050 |
0.1243004446 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 12 |
Hb_004393_030 |
0.1255445467 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 13 |
Hb_004109_150 |
0.1269542321 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 14 |
Hb_001821_050 |
0.1277021827 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 15 |
Hb_008202_010 |
0.1279921895 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000110_040 |
0.1283786634 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 17 |
Hb_134022_010 |
0.1284851719 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 18 |
Hb_001660_130 |
0.1304143212 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR4 [Jatropha curcas] |
| 19 |
Hb_010180_050 |
0.1315002765 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 20 |
Hb_008810_030 |
0.1322747856 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |