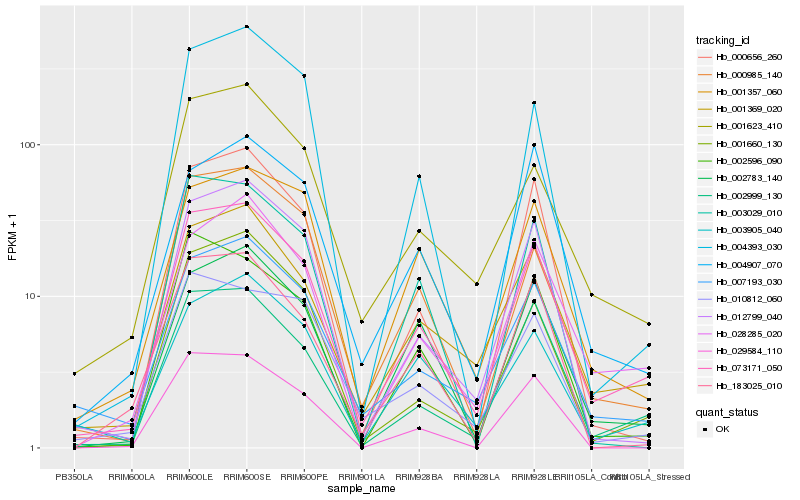
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073171_050 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 2 |
Hb_001369_020 |
0.0857344592 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 3 |
Hb_002999_130 |
0.0918318666 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 4 |
Hb_007193_030 |
0.0995623415 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 5 |
Hb_028285_020 |
0.1010963839 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_001623_410 |
0.1095265677 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_012799_040 |
0.1162723339 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 8 |
Hb_000985_140 |
0.1176087019 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 9 |
Hb_002783_140 |
0.1178334179 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 10 |
Hb_001660_130 |
0.1207271892 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR4 [Jatropha curcas] |
| 11 |
Hb_003905_040 |
0.1209638209 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 12 |
Hb_000656_260 |
0.1243004446 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 13 |
Hb_001357_060 |
0.1278095681 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 14 |
Hb_183025_010 |
0.1349191815 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 15 |
Hb_002596_090 |
0.1359748775 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_010812_060 |
0.1367410289 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 17 |
Hb_003029_010 |
0.1369267085 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004393_030 |
0.1370946631 |
- |
- |
CBL-interacting protein kinase 6 [Populus trichocarpa] |
| 19 |
Hb_029584_110 |
0.1399925441 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 20 |
Hb_004907_070 |
0.1400181773 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |