| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
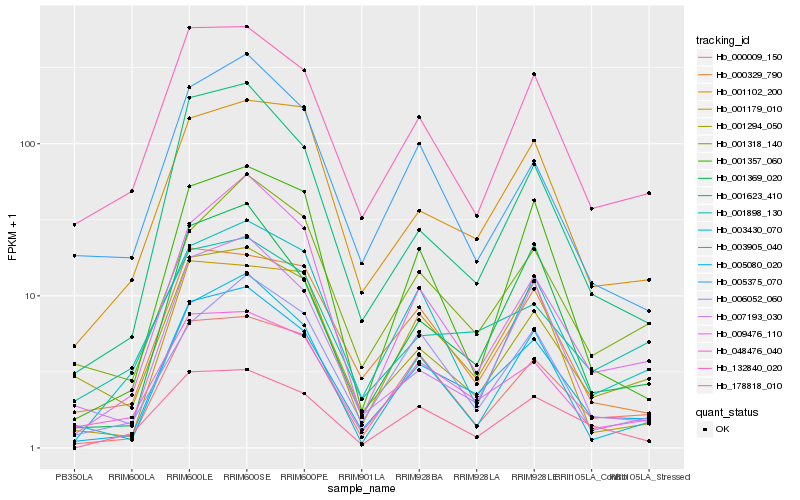
| 1 |
Hb_005080_020 |
0.0 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 2 |
Hb_048476_040 |
0.1056476833 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 3 |
Hb_001369_020 |
0.1075236293 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 4 |
Hb_132840_020 |
0.1083677772 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 5 |
Hb_001623_410 |
0.1127447762 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000009_150 |
0.1142618509 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 7 |
Hb_000329_790 |
0.1238400876 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 8 |
Hb_003905_040 |
0.1250167652 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 9 |
Hb_003430_070 |
0.1269332303 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 10 |
Hb_001102_200 |
0.1279080584 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 11 |
Hb_001294_050 |
0.1304169421 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 12 |
Hb_009476_110 |
0.1305361108 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 13 |
Hb_178818_010 |
0.130947669 |
- |
- |
CRINKLY4 related 3, putative [Theobroma cacao] |
| 14 |
Hb_001318_140 |
0.1335516147 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 15 |
Hb_007193_030 |
0.1338145975 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001357_060 |
0.1384494296 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 17 |
Hb_001179_010 |
0.139185494 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 18 |
Hb_006052_060 |
0.1393892865 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 19 |
Hb_005375_070 |
0.1401502089 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 20 |
Hb_001898_130 |
0.1406925691 |
- |
- |
ATP binding protein, putative [Ricinus communis] |