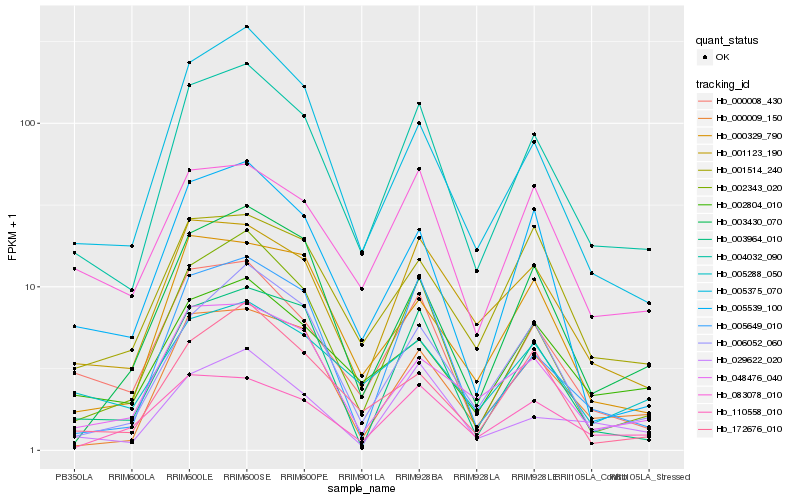
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_090 |
0.0 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 2 |
Hb_000008_430 |
0.1046817556 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002343_020 |
0.1125990741 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 4 |
Hb_001123_190 |
0.1132958154 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 5 |
Hb_000009_150 |
0.1149887002 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 6 |
Hb_048476_040 |
0.1252825531 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 7 |
Hb_006052_060 |
0.1272699076 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_002804_010 |
0.1290260815 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 9 |
Hb_029622_020 |
0.1317690988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_083078_010 |
0.1325476997 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 11 |
Hb_005375_070 |
0.1333256869 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 12 |
Hb_005288_050 |
0.1362749654 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 13 |
Hb_000329_790 |
0.1364187641 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 14 |
Hb_005649_010 |
0.1364545021 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 15 |
Hb_003964_010 |
0.1367970413 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 16 |
Hb_005539_100 |
0.1369079776 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 17 |
Hb_003430_070 |
0.1370056422 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 18 |
Hb_001514_240 |
0.1373666407 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 19 |
Hb_110558_010 |
0.1383766681 |
- |
- |
PREDICTED: uncharacterized protein LOC105641877 [Jatropha curcas] |
| 20 |
Hb_172676_010 |
0.1414899258 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |