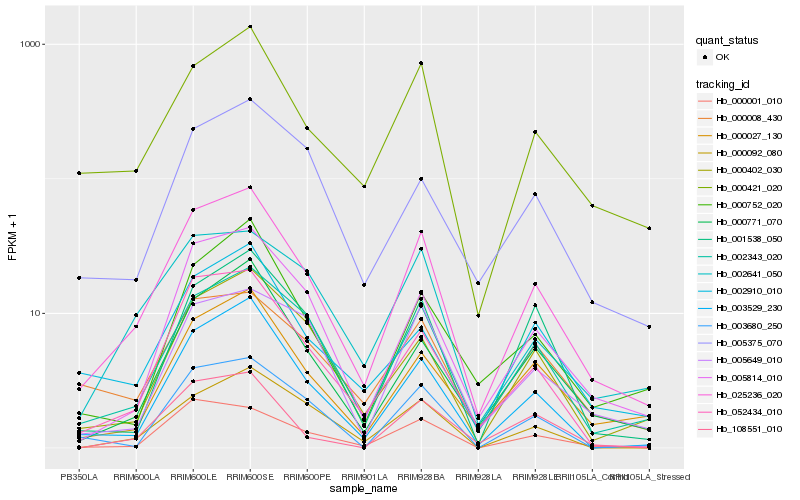
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025236_020 |
0.0 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 2 |
Hb_002343_020 |
0.1084178509 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 3 |
Hb_000421_020 |
0.120357944 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 4 |
Hb_005814_010 |
0.1205400944 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_052434_010 |
0.1206716376 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 6 |
Hb_003529_230 |
0.122330724 |
transcription factor |
TF Family: ERF |
- |
| 7 |
Hb_000771_070 |
0.1285725211 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000027_130 |
0.1289856453 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 9 |
Hb_000402_030 |
0.1410546992 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 10 |
Hb_002641_050 |
0.1454499447 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 11 |
Hb_000008_430 |
0.1475272614 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000001_010 |
0.1503892834 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |
| 13 |
Hb_002910_010 |
0.1509794902 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 14 |
Hb_000752_020 |
0.1540851909 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000092_080 |
0.15499845 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_038742 [Vitis vinifera] |
| 16 |
Hb_001538_050 |
0.1553309669 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 17 |
Hb_005375_070 |
0.1580163019 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 18 |
Hb_005649_010 |
0.1601096704 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 19 |
Hb_108551_010 |
0.1614322134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003680_250 |
0.1624985444 |
- |
- |
PREDICTED: probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial isoform X2 [Jatropha curcas] |