| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
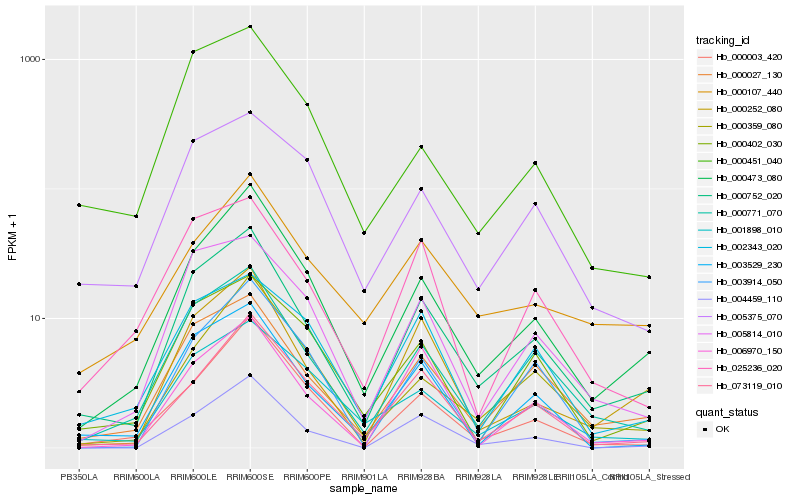
Hb_000752_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000027_130 |
0.0880960299 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 3 |
Hb_000771_070 |
0.0971664572 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000473_080 |
0.1047987817 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 5 |
Hb_003529_230 |
0.1307575312 |
transcription factor |
TF Family: ERF |
- |
| 6 |
Hb_000252_080 |
0.1316510045 |
- |
- |
PREDICTED: uncharacterized protein LOC105644644 [Jatropha curcas] |
| 7 |
Hb_005814_010 |
0.1359522206 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_001898_010 |
0.1427630209 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 9 |
Hb_004459_110 |
0.1468772051 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 10 |
Hb_073119_010 |
0.1486320829 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 11 |
Hb_000402_030 |
0.1535374781 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 12 |
Hb_025236_020 |
0.1540851909 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 13 |
Hb_000359_080 |
0.1548698224 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 14 |
Hb_000003_420 |
0.1568648656 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_005375_070 |
0.1595627816 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 16 |
Hb_000107_440 |
0.1601101134 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 17 |
Hb_002343_020 |
0.1617148029 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 18 |
Hb_006970_150 |
0.1651548445 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 19 |
Hb_000451_040 |
0.1663231667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003914_050 |
0.166973743 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |