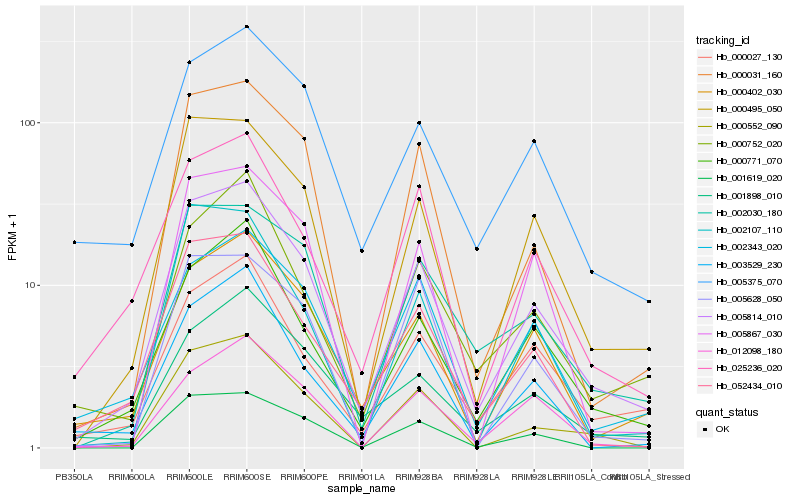
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005814_010 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000495_050 |
0.0842184606 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 3 |
Hb_000771_070 |
0.1072310561 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000031_160 |
0.1116906564 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 5 |
Hb_000402_030 |
0.1130908255 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 6 |
Hb_052434_010 |
0.1152593159 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 7 |
Hb_000552_090 |
0.117188627 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 8 |
Hb_002030_180 |
0.1193882053 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_000027_130 |
0.1197752568 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 10 |
Hb_025236_020 |
0.1205400944 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 11 |
Hb_005375_070 |
0.1280448022 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 12 |
Hb_002343_020 |
0.1280981274 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 13 |
Hb_001898_010 |
0.1308407406 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 14 |
Hb_003529_230 |
0.1330952423 |
transcription factor |
TF Family: ERF |
- |
| 15 |
Hb_001619_020 |
0.1344076584 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 16 |
Hb_012098_180 |
0.1345960769 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 17 |
Hb_002107_110 |
0.1355690163 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000752_020 |
0.1359522206 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005867_030 |
0.1381137329 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 20 |
Hb_005628_050 |
0.1458976623 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |