| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
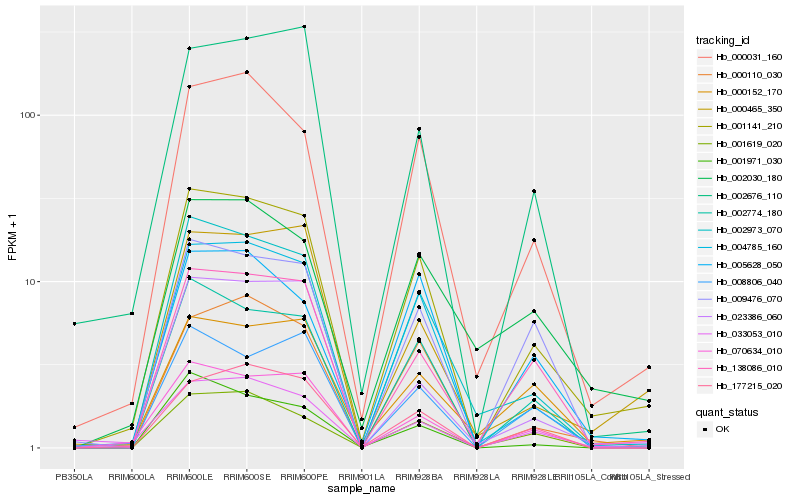
Hb_001141_210 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 2 |
Hb_004785_160 |
0.1002152111 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 3 |
Hb_002774_180 |
0.1035549976 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_000031_160 |
0.10690173 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 5 |
Hb_002973_070 |
0.1110015572 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 6 |
Hb_138086_010 |
0.1157000794 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 7 |
Hb_070634_010 |
0.1214977011 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 8 |
Hb_000110_030 |
0.1253382247 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_177215_020 |
0.1356136372 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 10 |
Hb_000465_350 |
0.135679862 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2 [Jatropha curcas] |
| 11 |
Hb_033053_010 |
0.139479148 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_001619_020 |
0.1418409563 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 13 |
Hb_000152_170 |
0.143864562 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 14 |
Hb_005628_050 |
0.1443689435 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_023386_060 |
0.1444191985 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 16 |
Hb_008806_040 |
0.1447753013 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 17 |
Hb_009476_070 |
0.1459312281 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 18 |
Hb_001971_030 |
0.1484791582 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 19 |
Hb_002030_180 |
0.1514541348 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_002676_110 |
0.1524261489 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |