| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
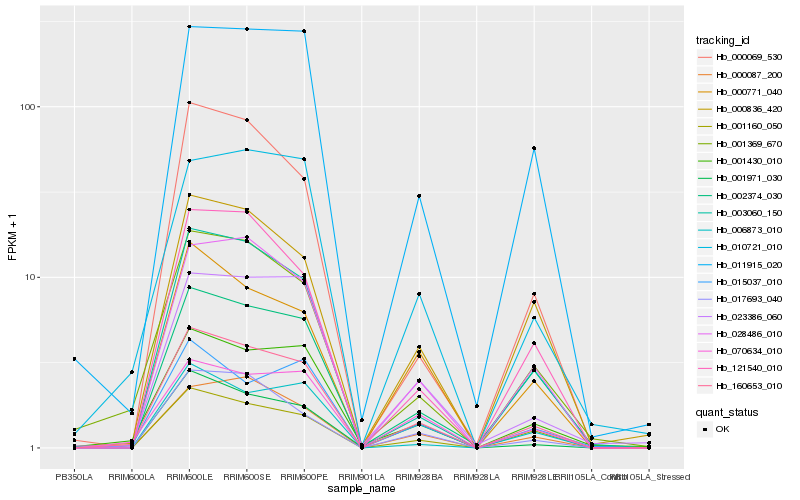
Hb_160653_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 2 |
Hb_028486_010 |
0.1014906312 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 3 |
Hb_001430_010 |
0.1020695132 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003060_150 |
0.1038275958 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_001369_670 |
0.1093445511 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 6 |
Hb_001160_050 |
0.1103841907 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 7 |
Hb_070634_010 |
0.1130028383 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 8 |
Hb_000836_420 |
0.1178692645 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000771_040 |
0.1179720061 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Jatropha curcas] |
| 10 |
Hb_121540_010 |
0.1181149079 |
- |
- |
PREDICTED: UDP-glycosyltransferase 88A1-like [Jatropha curcas] |
| 11 |
Hb_000087_200 |
0.1184966691 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 12 |
Hb_011915_020 |
0.1245308073 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 6, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000069_530 |
0.1246038261 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 14 |
Hb_002374_030 |
0.1250429742 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 15 |
Hb_017693_040 |
0.1259434899 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 16 |
Hb_023386_060 |
0.1275529036 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 17 |
Hb_010721_010 |
0.1278903229 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 18 |
Hb_015037_010 |
0.1316127697 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 19 |
Hb_001971_030 |
0.1326420653 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 20 |
Hb_006873_010 |
0.1343020735 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |