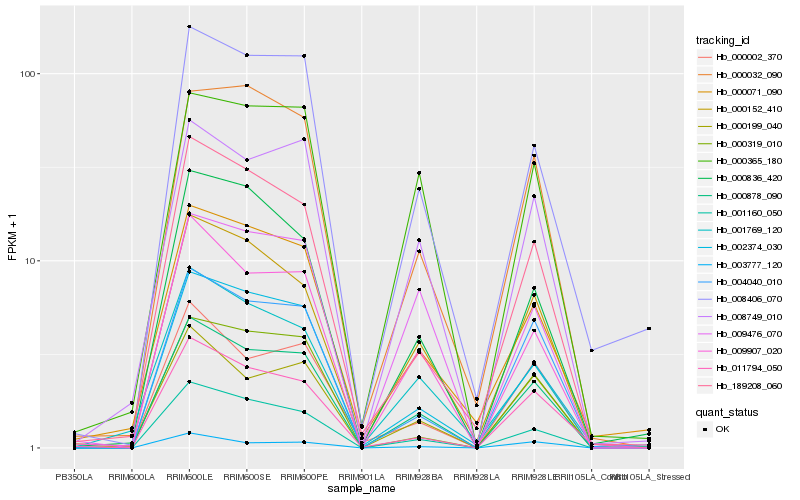
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000878_090 |
0.0 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 2 |
Hb_009907_020 |
0.0747840799 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 3 |
Hb_001160_050 |
0.0763574668 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 4 |
Hb_008749_010 |
0.0895128666 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 5 |
Hb_002374_030 |
0.0941000092 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004040_010 |
0.0944083517 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 7 |
Hb_000152_410 |
0.0951082789 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 8 |
Hb_000199_040 |
0.0996167284 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000002_370 |
0.1064693533 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 10 |
Hb_008406_070 |
0.1088380277 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 11 |
Hb_189208_060 |
0.1095117656 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 12 |
Hb_011794_050 |
0.1127569452 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000319_010 |
0.1131271643 |
- |
- |
hypothetical protein POPTR_0016s03600g [Populus trichocarpa] |
| 14 |
Hb_000032_090 |
0.1152741427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000836_420 |
0.1185997382 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 16 |
Hb_009476_070 |
0.1196370691 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 17 |
Hb_001769_120 |
0.1211045064 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 18 |
Hb_000365_180 |
0.1246136448 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 19 |
Hb_003777_120 |
0.124907673 |
- |
- |
- |
| 20 |
Hb_000071_090 |
0.1253040324 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |