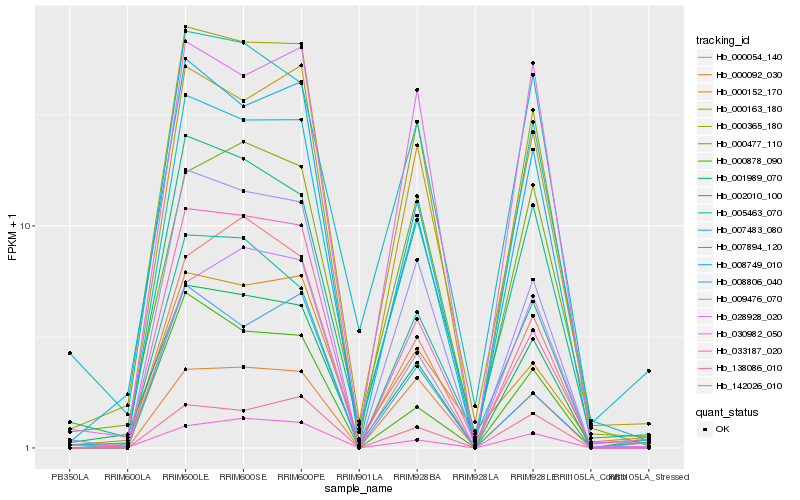
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_180 |
0.0 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 2 |
Hb_009476_070 |
0.0691036828 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 3 |
Hb_000163_180 |
0.0778209089 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 4 |
Hb_001989_070 |
0.0790595532 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 5 |
Hb_008749_010 |
0.0793886534 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 6 |
Hb_002010_100 |
0.0988419811 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 7 |
Hb_033187_020 |
0.1047508933 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 8 |
Hb_138086_010 |
0.1048964783 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 9 |
Hb_005463_070 |
0.1105118907 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_142026_010 |
0.112492605 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 11 |
Hb_007894_120 |
0.1152240419 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000152_170 |
0.1155990843 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 13 |
Hb_028928_020 |
0.1185995516 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 14 |
Hb_000054_140 |
0.1221631942 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 15 |
Hb_000092_030 |
0.12308136 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 16 |
Hb_000477_110 |
0.1244418687 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 17 |
Hb_000878_090 |
0.1246136448 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 18 |
Hb_030982_050 |
0.1264491275 |
- |
- |
PREDICTED: gamma-glutamyl hydrolase 2-like [Jatropha curcas] |
| 19 |
Hb_008806_040 |
0.1271923831 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 20 |
Hb_007483_080 |
0.1283447812 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |