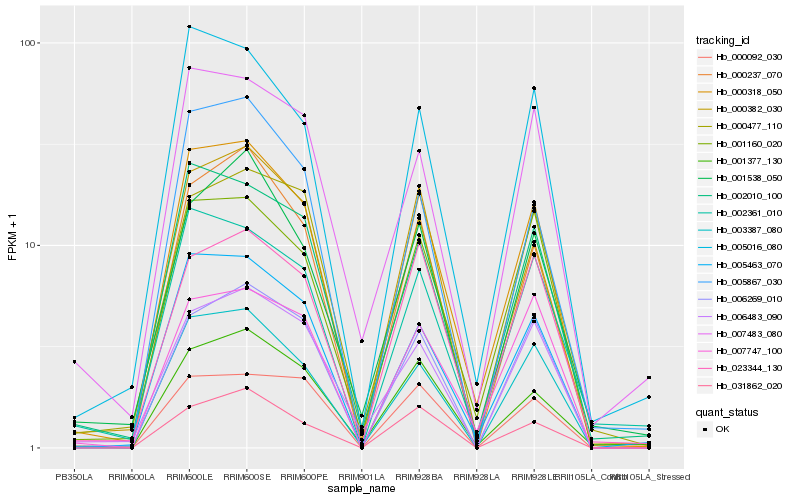
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_050 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005463_070 |
0.0762225539 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_003387_080 |
0.0863230648 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000382_030 |
0.0880407144 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 5 |
Hb_001160_020 |
0.0891969206 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 6 |
Hb_005016_080 |
0.0920964841 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 7 |
Hb_002010_100 |
0.0921237069 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 8 |
Hb_000237_070 |
0.0946310625 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 9 |
Hb_001377_130 |
0.1074956895 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 10 |
Hb_002361_010 |
0.1085158158 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_031862_020 |
0.1125612555 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 12 |
Hb_005867_030 |
0.1129713633 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 13 |
Hb_007747_100 |
0.1130383968 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 14 |
Hb_006269_010 |
0.1131051358 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 15 |
Hb_023344_130 |
0.1197063619 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_000477_110 |
0.1200327829 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 17 |
Hb_007483_080 |
0.1211586345 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 18 |
Hb_006483_090 |
0.1238780224 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 19 |
Hb_000092_030 |
0.1243090605 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 20 |
Hb_001538_050 |
0.1259170008 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |