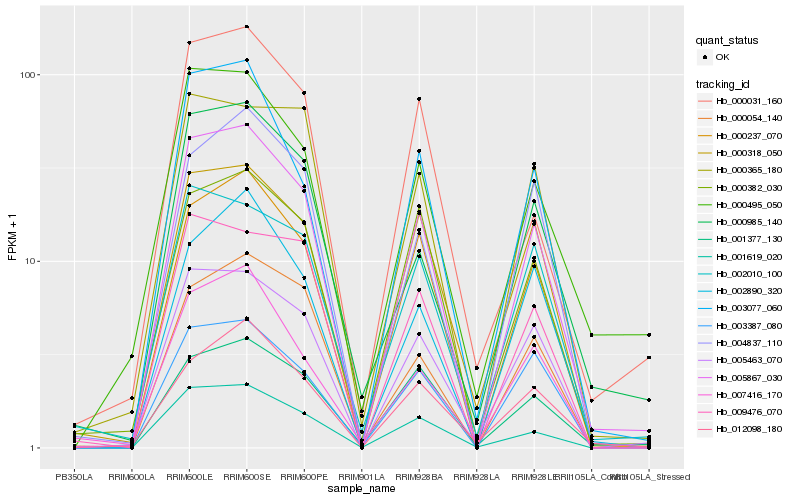
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005867_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 2 |
Hb_000237_070 |
0.0835578296 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 3 |
Hb_005463_070 |
0.0855125633 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_001619_020 |
0.0856854386 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 5 |
Hb_003077_060 |
0.1031598581 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 6 |
Hb_000495_050 |
0.1128225376 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 7 |
Hb_000318_050 |
0.1129713633 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000054_140 |
0.1144440284 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 9 |
Hb_009476_070 |
0.1160500281 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 10 |
Hb_003387_080 |
0.1161453859 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000985_140 |
0.1183130723 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 12 |
Hb_004837_110 |
0.1195671777 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000382_030 |
0.1197035642 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 14 |
Hb_012098_180 |
0.1223453919 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 15 |
Hb_002010_100 |
0.1228866425 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 16 |
Hb_002890_320 |
0.1247204655 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 17 |
Hb_001377_130 |
0.1252392318 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 18 |
Hb_000031_160 |
0.1285631254 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 19 |
Hb_000365_180 |
0.1302711954 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 20 |
Hb_007416_170 |
0.1310017751 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |