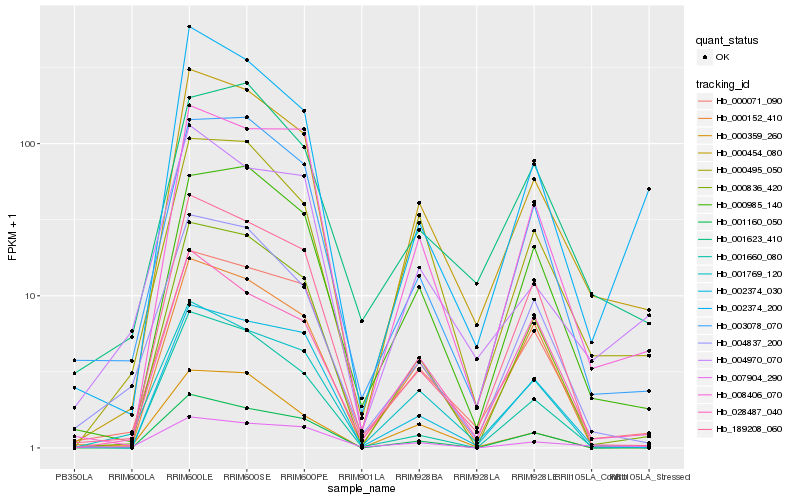
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000454_080 |
0.0 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 2 |
Hb_000071_090 |
0.1022769049 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 3 |
Hb_000836_420 |
0.1037506798 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000495_050 |
0.1123333948 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 5 |
Hb_000985_140 |
0.1159104701 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 6 |
Hb_008406_070 |
0.1192336171 |
- |
- |
PREDICTED: probable protein phosphatase 2C 58 [Jatropha curcas] |
| 7 |
Hb_001769_120 |
0.1280330953 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 8 |
Hb_003078_070 |
0.1280952159 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g25290 [Jatropha curcas] |
| 9 |
Hb_000152_410 |
0.1301122271 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 10 |
Hb_002374_200 |
0.1312547066 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 11 |
Hb_001160_050 |
0.1330155286 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 12 |
Hb_002374_030 |
0.1331054263 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 13 |
Hb_189208_060 |
0.1339802082 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 14 |
Hb_028487_040 |
0.1343751947 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |
| 15 |
Hb_001623_410 |
0.1397643009 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004837_200 |
0.1405018024 |
- |
- |
hypothetical protein POPTR_0001s26450g [Populus trichocarpa] |
| 17 |
Hb_001660_080 |
0.1409719613 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 18 |
Hb_004970_070 |
0.1420310506 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 19 |
Hb_007904_290 |
0.142956964 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 20 |
Hb_000359_260 |
0.1463199433 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |