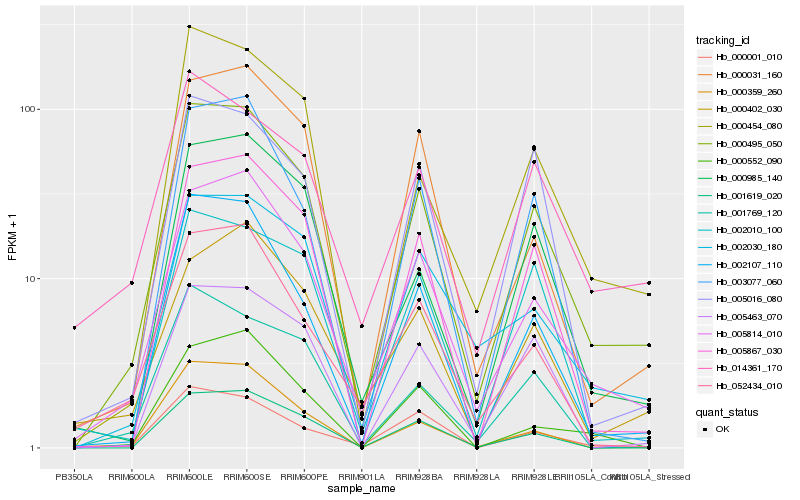
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000495_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 2 |
Hb_005814_010 |
0.0842184606 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_002107_110 |
0.1083867329 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000454_080 |
0.1123333948 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 5 |
Hb_005867_030 |
0.1128225376 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 6 |
Hb_000985_140 |
0.1245814577 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 7 |
Hb_002030_180 |
0.1272800482 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_001619_020 |
0.1275113541 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 9 |
Hb_005463_070 |
0.1275126525 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_003077_060 |
0.133171819 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 11 |
Hb_000031_160 |
0.1343925226 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 12 |
Hb_000359_260 |
0.1352603377 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 13 |
Hb_001769_120 |
0.1411427434 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 14 |
Hb_005016_080 |
0.142033687 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 15 |
Hb_014361_170 |
0.1442459249 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |
| 16 |
Hb_000402_030 |
0.1454687559 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 17 |
Hb_002010_100 |
0.1460188209 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 18 |
Hb_000552_090 |
0.1471257421 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 19 |
Hb_052434_010 |
0.1475807889 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 20 |
Hb_000001_010 |
0.1476655175 |
- |
- |
PREDICTED: uncharacterized protein LOC105648961 [Jatropha curcas] |