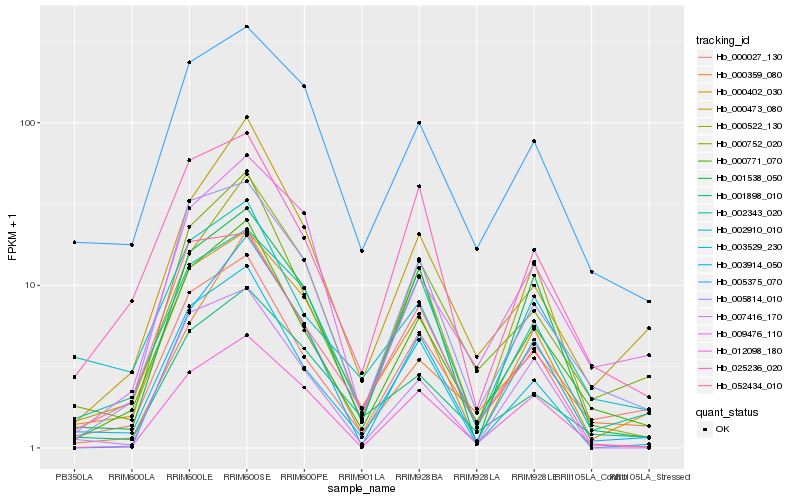
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000771_070 |
0.0 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000027_130 |
0.0717595158 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 3 |
Hb_000752_020 |
0.0971664572 |
- |
- |
PREDICTED: uncharacterized protein LOC105641803 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005814_010 |
0.1072310561 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_003529_230 |
0.1248565021 |
transcription factor |
TF Family: ERF |
- |
| 6 |
Hb_000402_030 |
0.1269357941 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 7 |
Hb_025236_020 |
0.1285725211 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 8 |
Hb_000473_080 |
0.1298100252 |
- |
- |
PREDICTED: lipase-like PAD4 [Populus euphratica] |
| 9 |
Hb_012098_180 |
0.1335887535 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 10 |
Hb_002910_010 |
0.1355724208 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 11 |
Hb_000522_130 |
0.1358875793 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 12 |
Hb_001898_010 |
0.1362011849 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 13 |
Hb_005375_070 |
0.1409083345 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 14 |
Hb_009476_110 |
0.1433160816 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 15 |
Hb_003914_050 |
0.1457948934 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 16 |
Hb_000359_080 |
0.1467280961 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 17 |
Hb_001538_050 |
0.1488277783 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 18 |
Hb_002343_020 |
0.150489805 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 19 |
Hb_007416_170 |
0.151400309 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 20 |
Hb_052434_010 |
0.1520097167 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |