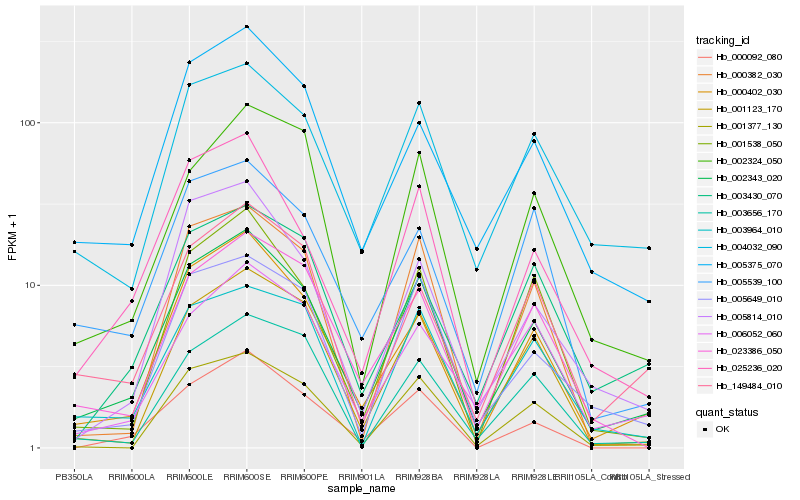
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002343_020 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 2 |
Hb_000402_030 |
0.0912925629 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 3 |
Hb_025236_020 |
0.1084178509 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 4 |
Hb_005649_010 |
0.1088076153 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 5 |
Hb_004032_090 |
0.1125990741 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 6 |
Hb_001123_170 |
0.1142018649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005375_070 |
0.1152201857 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 8 |
Hb_003964_010 |
0.1210162442 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 9 |
Hb_001538_050 |
0.1213339851 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 10 |
Hb_001377_130 |
0.1213595927 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 11 |
Hb_149484_010 |
0.1223052947 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 12 |
Hb_023386_050 |
0.1261079242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006052_060 |
0.1262688959 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 14 |
Hb_005814_010 |
0.1280981274 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_002324_050 |
0.129525599 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 16 |
Hb_000092_080 |
0.1331208719 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_038742 [Vitis vinifera] |
| 17 |
Hb_003656_170 |
0.1342245276 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 18 |
Hb_000382_030 |
0.1348817351 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 19 |
Hb_003430_070 |
0.1352194185 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 20 |
Hb_005539_100 |
0.1355996619 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |