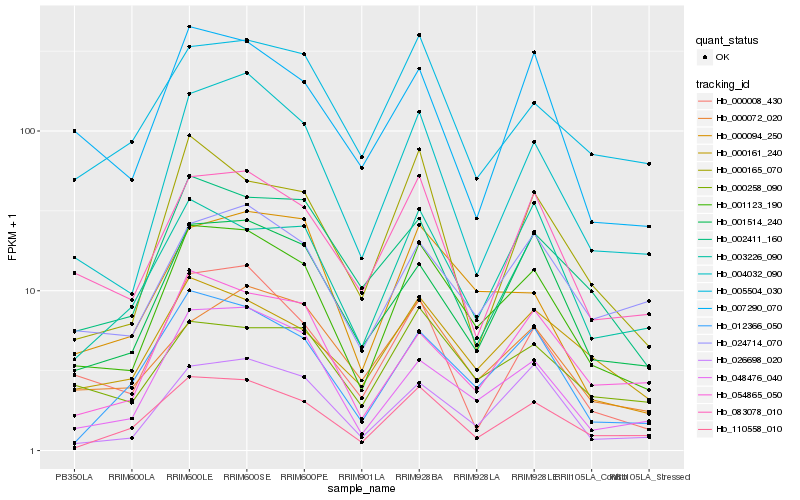
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_190 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 2 |
Hb_004032_090 |
0.1132958154 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 3 |
Hb_110558_010 |
0.1161461938 |
- |
- |
PREDICTED: uncharacterized protein LOC105641877 [Jatropha curcas] |
| 4 |
Hb_048476_040 |
0.1187073569 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 5 |
Hb_012366_050 |
0.120884791 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 6 |
Hb_026698_020 |
0.1274681479 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 7 |
Hb_083078_010 |
0.1276600357 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 8 |
Hb_001514_240 |
0.1286166733 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 9 |
Hb_000161_240 |
0.1305543503 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_054865_050 |
0.1381354335 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_024714_070 |
0.1390567492 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 12 |
Hb_000008_430 |
0.1402862096 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 24 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000258_090 |
0.1405797373 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000094_250 |
0.1432140502 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X3 [Jatropha curcas] |
| 15 |
Hb_002411_160 |
0.1447662148 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007290_070 |
0.1448428462 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 17 |
Hb_003226_090 |
0.1453754525 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 18 |
Hb_000165_070 |
0.146300286 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 19 |
Hb_005504_030 |
0.1463546287 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 20 |
Hb_000072_020 |
0.1464809726 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |