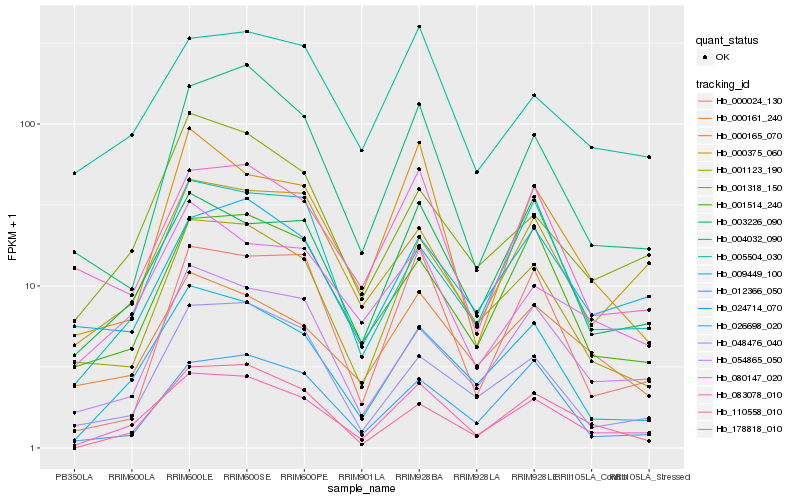
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_110558_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641877 [Jatropha curcas] |
| 2 |
Hb_012366_050 |
0.0993765445 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 3 |
Hb_001123_190 |
0.1161461938 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 4 |
Hb_003226_090 |
0.1309900978 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 5 |
Hb_005504_030 |
0.1323434672 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 6 |
Hb_054865_050 |
0.1360655884 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 7 |
Hb_004032_090 |
0.1383766681 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 8 |
Hb_001514_240 |
0.1396252235 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 9 |
Hb_001318_150 |
0.142721342 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 10 |
Hb_000161_240 |
0.1434178795 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_178818_010 |
0.144948212 |
- |
- |
CRINKLY4 related 3, putative [Theobroma cacao] |
| 12 |
Hb_083078_010 |
0.1449708155 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 13 |
Hb_080147_020 |
0.1451237471 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 14 |
Hb_000165_070 |
0.1458590739 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 15 |
Hb_026698_020 |
0.1460053142 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 16 |
Hb_024714_070 |
0.146643669 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 17 |
Hb_009449_100 |
0.1473630924 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 18 |
Hb_000375_060 |
0.147649561 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 19 |
Hb_000024_130 |
0.1510400954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_048476_040 |
0.1515292576 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |