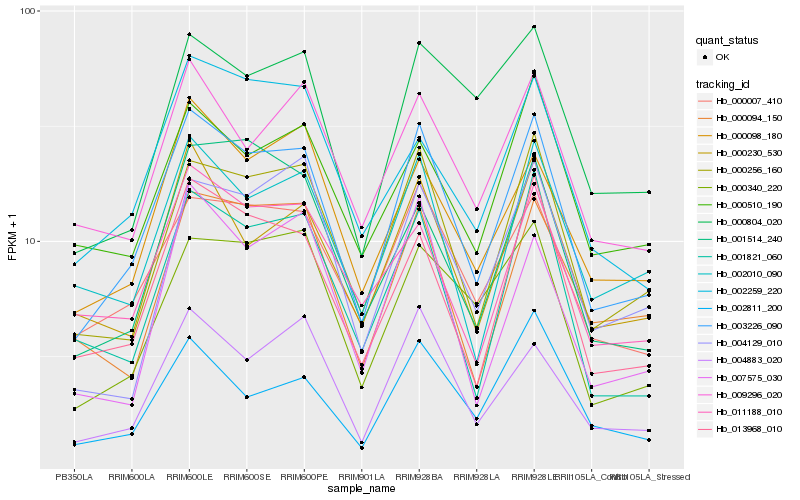
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003226_090 |
0.0 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 2 |
Hb_000256_160 |
0.083414535 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 3 |
Hb_013968_010 |
0.0952884441 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 4 |
Hb_000007_410 |
0.097245471 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 5 |
Hb_004883_020 |
0.1035490681 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 6 |
Hb_002010_090 |
0.1044065536 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000230_530 |
0.1076962118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001821_060 |
0.1091369231 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 9 |
Hb_000340_220 |
0.1129063815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009296_020 |
0.113513029 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_000098_180 |
0.1141498838 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000094_150 |
0.1163592795 |
- |
- |
- |
| 13 |
Hb_011188_010 |
0.1190474151 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000510_190 |
0.1231867333 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 15 |
Hb_002811_200 |
0.12346401 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 16 |
Hb_002259_220 |
0.1239085897 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 17 |
Hb_007575_030 |
0.1239868852 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 18 |
Hb_004129_010 |
0.1243131515 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 19 |
Hb_000804_020 |
0.1246913845 |
- |
- |
PREDICTED: aconitate hydratase, cytoplasmic [Jatropha curcas] |
| 20 |
Hb_001514_240 |
0.1267359756 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |