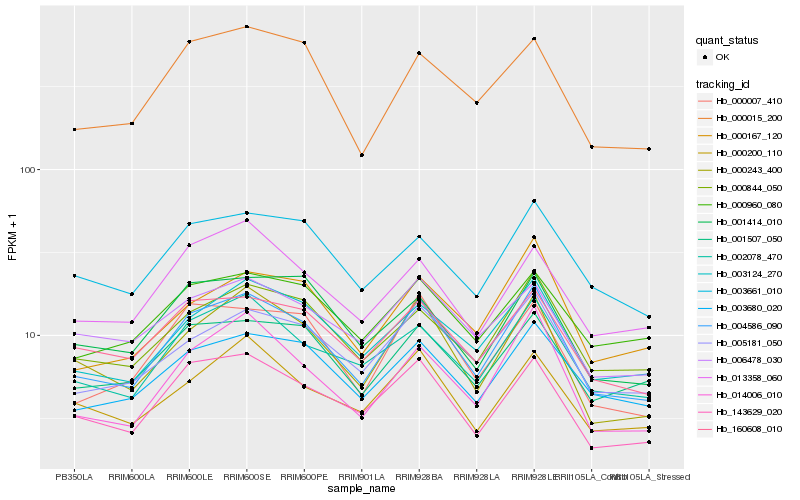
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_090 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 2 |
Hb_143629_020 |
0.0655930947 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 3 |
Hb_000243_400 |
0.0704289987 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 4 |
Hb_003124_270 |
0.0725977857 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 5 |
Hb_005181_050 |
0.0738624837 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 6 |
Hb_002078_470 |
0.075486565 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000844_050 |
0.0767074916 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 8 |
Hb_160608_010 |
0.0769983449 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 9 |
Hb_003680_020 |
0.0781006274 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_014006_010 |
0.0786682217 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 11 |
Hb_000007_410 |
0.0803726201 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 12 |
Hb_001507_050 |
0.0839468533 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000015_200 |
0.0853436352 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 14 |
Hb_000200_110 |
0.0875479638 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003661_010 |
0.0880581057 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000167_120 |
0.0911970896 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 17 |
Hb_006478_030 |
0.0912366487 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 18 |
Hb_013358_060 |
0.0922477643 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 19 |
Hb_001414_010 |
0.0934270106 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 20 |
Hb_000960_080 |
0.093746351 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |