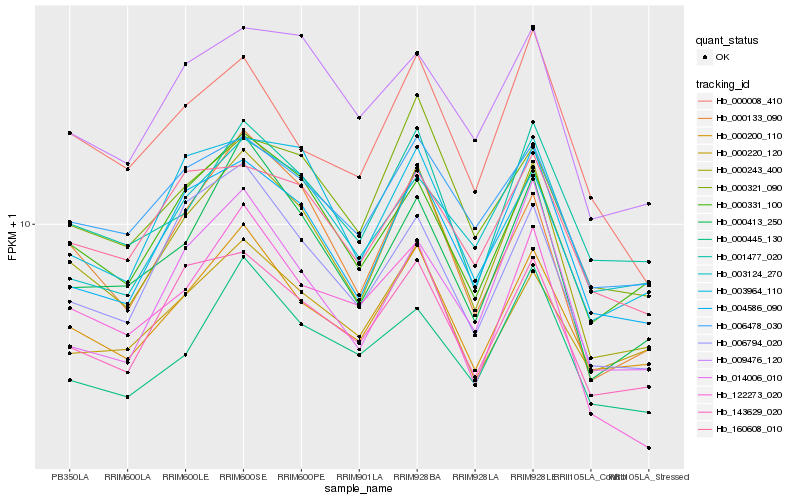
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_400 |
0.0 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 2 |
Hb_004586_090 |
0.0704289987 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000133_090 |
0.0763741501 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 4 |
Hb_000200_110 |
0.0764865866 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000413_250 |
0.0780152208 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 6 |
Hb_143629_020 |
0.082004231 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 7 |
Hb_006794_020 |
0.0878014435 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 8 |
Hb_000331_100 |
0.0880031443 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001477_020 |
0.0977229536 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 10 |
Hb_014006_010 |
0.1011743551 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 11 |
Hb_006478_030 |
0.1034973337 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 12 |
Hb_009476_120 |
0.1038839713 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 13 |
Hb_003964_110 |
0.1052117048 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000321_090 |
0.1056041016 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 15 |
Hb_000445_130 |
0.1057974232 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 16 |
Hb_003124_270 |
0.106036316 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 17 |
Hb_160608_010 |
0.1075426333 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 18 |
Hb_122273_020 |
0.1086349929 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP2 [Jatropha curcas] |
| 19 |
Hb_000220_120 |
0.1087734132 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000008_410 |
0.1094960766 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |