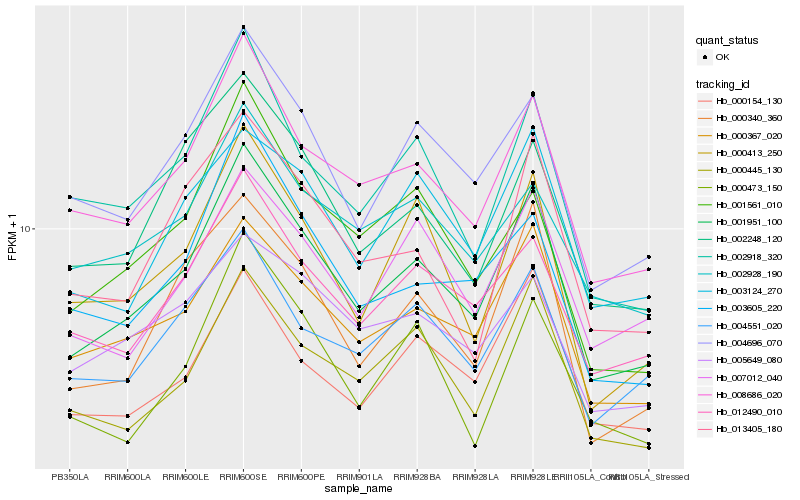
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_100 |
0.0 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 2 |
Hb_008686_020 |
0.0659756989 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000367_020 |
0.0682331175 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 4 |
Hb_004696_070 |
0.0793402352 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 5 |
Hb_012490_010 |
0.081706592 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000445_130 |
0.0824001319 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 7 |
Hb_000340_360 |
0.0866105264 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 8 |
Hb_000413_250 |
0.0867114374 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 9 |
Hb_002918_320 |
0.0869653091 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004551_020 |
0.0877795592 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 11 |
Hb_000154_130 |
0.0888260848 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 12 |
Hb_007012_040 |
0.0919140728 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 13 |
Hb_002248_120 |
0.0926381438 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 14 |
Hb_003605_220 |
0.0945265173 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 15 |
Hb_003124_270 |
0.0950478697 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 16 |
Hb_013405_180 |
0.0973592922 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 17 |
Hb_001561_010 |
0.0983896362 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005649_080 |
0.0985542088 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 19 |
Hb_002928_190 |
0.0993290135 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 20 |
Hb_000473_150 |
0.1052871115 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |