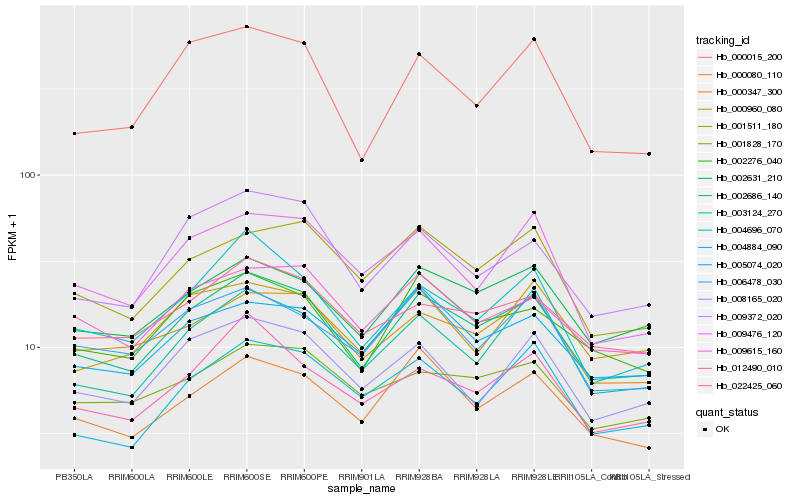
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_140 |
0.0 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002631_210 |
0.0648659577 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 3 |
Hb_002276_040 |
0.0686384029 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 4 |
Hb_000080_110 |
0.0745491656 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000347_300 |
0.074882169 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 6 |
Hb_009615_160 |
0.0770070616 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 7 |
Hb_022425_060 |
0.0776039794 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_004696_070 |
0.078336006 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 9 |
Hb_003124_270 |
0.0783859014 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 10 |
Hb_005074_020 |
0.078839432 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001511_180 |
0.079794736 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 12 |
Hb_008165_020 |
0.080406486 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000015_200 |
0.0804910691 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 14 |
Hb_009372_020 |
0.0805798644 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 15 |
Hb_006478_030 |
0.081828341 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 16 |
Hb_012490_010 |
0.0825987147 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001828_170 |
0.0828743658 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 18 |
Hb_009476_120 |
0.0873795159 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 19 |
Hb_004884_090 |
0.0894482947 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 20 |
Hb_000960_080 |
0.0904361361 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |