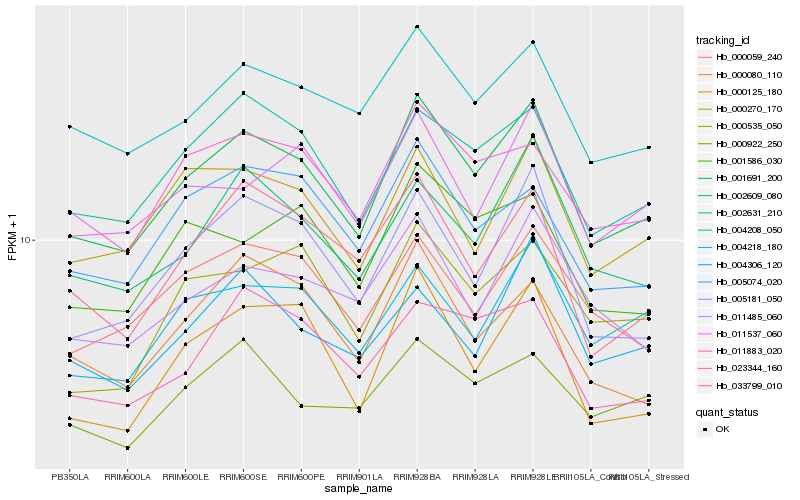
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001691_200 |
0.0 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 2 |
Hb_002631_210 |
0.0668608426 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 3 |
Hb_004208_050 |
0.0713338758 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 4 |
Hb_011883_020 |
0.0740457037 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 5 |
Hb_002609_080 |
0.0758622205 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_005181_050 |
0.0785342355 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000270_170 |
0.0812700916 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 8 |
Hb_000059_240 |
0.0815909307 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 9 |
Hb_004218_180 |
0.0823296964 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 10 |
Hb_005074_020 |
0.083216287 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 11 |
Hb_011485_060 |
0.0852590772 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 12 |
Hb_011537_060 |
0.0859510673 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 13 |
Hb_001586_030 |
0.0870372578 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 14 |
Hb_000922_250 |
0.0873026379 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_033799_010 |
0.0873114396 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 16 |
Hb_023344_160 |
0.0874383583 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 17 |
Hb_000535_050 |
0.0898290959 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 18 |
Hb_000125_180 |
0.0902294879 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 19 |
Hb_000080_110 |
0.0911820691 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004306_120 |
0.0923991602 |
- |
- |
DNA binding protein, putative [Ricinus communis] |